
Wuhan spider virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
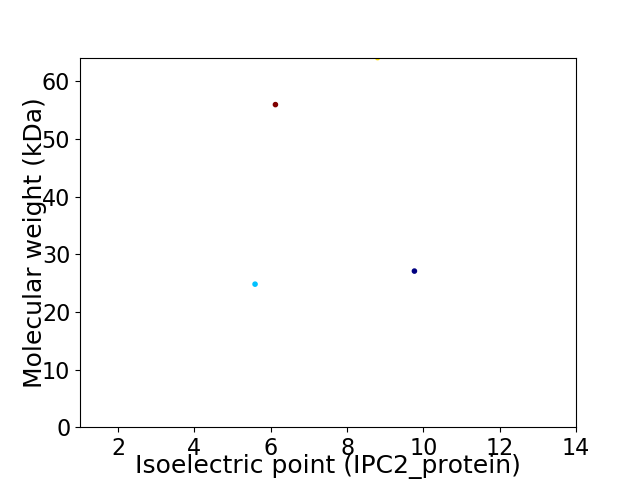
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
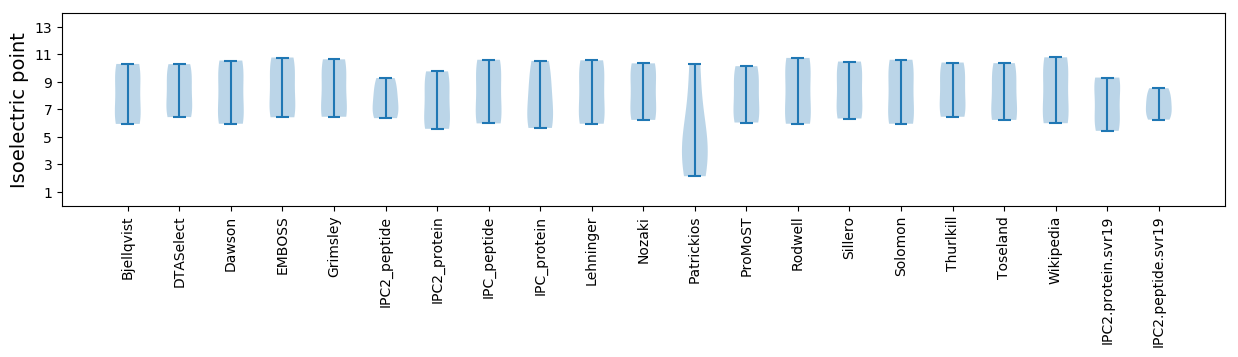
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG33|A0A1L3KG33_9VIRU RNA-directed RNA polymerase OS=Wuhan spider virus 9 OX=1923758 PE=4 SV=1
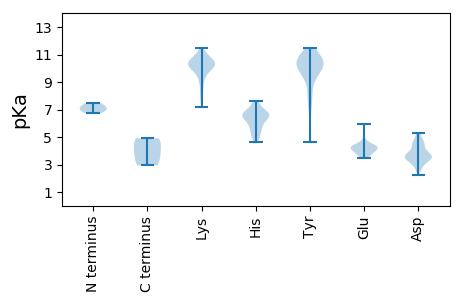
MM1 pKa = 7.14HH2 pKa = 7.62AIVVTDD8 pKa = 5.32HH9 pKa = 6.32IWTNYY14 pKa = 8.17GRR16 pKa = 11.84YY17 pKa = 9.18AATAQSFINQQRR29 pKa = 11.84ALYY32 pKa = 9.98DD33 pKa = 3.76PNDD36 pKa = 4.39RR37 pKa = 11.84AWHH40 pKa = 6.53IAPQHH45 pKa = 5.28QPLAIVAHH53 pKa = 6.6DD54 pKa = 3.94WQWDD58 pKa = 3.39ATYY61 pKa = 10.51RR62 pKa = 11.84CYY64 pKa = 10.55ISNLDD69 pKa = 3.59YY70 pKa = 11.21SWAVDD75 pKa = 3.72YY76 pKa = 7.71THH78 pKa = 7.52KK79 pKa = 10.46ISPQFWFSVHH89 pKa = 4.94MPKK92 pKa = 10.43GIAMPVVGAVGSDD105 pKa = 2.72WDD107 pKa = 4.66VIQHH111 pKa = 7.0WIHH114 pKa = 5.95GVPVITHH121 pKa = 6.66IEE123 pKa = 4.1DD124 pKa = 3.35QVSRR128 pKa = 11.84GAKK131 pKa = 9.42IGGVEE136 pKa = 4.0YY137 pKa = 10.57NYY139 pKa = 10.81TDD141 pKa = 4.56LSINLQLTIPKK152 pKa = 9.62HH153 pKa = 4.61SQNFRR158 pKa = 11.84INLPLITLYY167 pKa = 10.33PDD169 pKa = 3.02TPYY172 pKa = 10.5IITVAATFYY181 pKa = 10.49MFTLEE186 pKa = 4.1RR187 pKa = 11.84AKK189 pKa = 10.6KK190 pKa = 8.11EE191 pKa = 3.83QPGFYY196 pKa = 10.41NLAEE200 pKa = 3.76VWFYY204 pKa = 11.39QEE206 pKa = 4.84GGAPDD211 pKa = 3.59EE212 pKa = 4.47PSRR215 pKa = 4.48
MM1 pKa = 7.14HH2 pKa = 7.62AIVVTDD8 pKa = 5.32HH9 pKa = 6.32IWTNYY14 pKa = 8.17GRR16 pKa = 11.84YY17 pKa = 9.18AATAQSFINQQRR29 pKa = 11.84ALYY32 pKa = 9.98DD33 pKa = 3.76PNDD36 pKa = 4.39RR37 pKa = 11.84AWHH40 pKa = 6.53IAPQHH45 pKa = 5.28QPLAIVAHH53 pKa = 6.6DD54 pKa = 3.94WQWDD58 pKa = 3.39ATYY61 pKa = 10.51RR62 pKa = 11.84CYY64 pKa = 10.55ISNLDD69 pKa = 3.59YY70 pKa = 11.21SWAVDD75 pKa = 3.72YY76 pKa = 7.71THH78 pKa = 7.52KK79 pKa = 10.46ISPQFWFSVHH89 pKa = 4.94MPKK92 pKa = 10.43GIAMPVVGAVGSDD105 pKa = 2.72WDD107 pKa = 4.66VIQHH111 pKa = 7.0WIHH114 pKa = 5.95GVPVITHH121 pKa = 6.66IEE123 pKa = 4.1DD124 pKa = 3.35QVSRR128 pKa = 11.84GAKK131 pKa = 9.42IGGVEE136 pKa = 4.0YY137 pKa = 10.57NYY139 pKa = 10.81TDD141 pKa = 4.56LSINLQLTIPKK152 pKa = 9.62HH153 pKa = 4.61SQNFRR158 pKa = 11.84INLPLITLYY167 pKa = 10.33PDD169 pKa = 3.02TPYY172 pKa = 10.5IITVAATFYY181 pKa = 10.49MFTLEE186 pKa = 4.1RR187 pKa = 11.84AKK189 pKa = 10.6KK190 pKa = 8.11EE191 pKa = 3.83QPGFYY196 pKa = 10.41NLAEE200 pKa = 3.76VWFYY204 pKa = 11.39QEE206 pKa = 4.84GGAPDD211 pKa = 3.59EE212 pKa = 4.47PSRR215 pKa = 4.48
Molecular weight: 24.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGI4|A0A1L3KGI4_9VIRU Uncharacterized protein OS=Wuhan spider virus 9 OX=1923758 PE=4 SV=1
MM1 pKa = 7.05MNQYY5 pKa = 10.25RR6 pKa = 11.84GRR8 pKa = 11.84GRR10 pKa = 11.84FWNAGLNLAAAAGGAAARR28 pKa = 11.84VAANRR33 pKa = 11.84FQNWVNSPPSNPYY46 pKa = 10.44NMQPIYY52 pKa = 10.27QALPPVRR59 pKa = 11.84RR60 pKa = 11.84GRR62 pKa = 11.84GRR64 pKa = 11.84GRR66 pKa = 11.84GRR68 pKa = 11.84GRR70 pKa = 11.84GRR72 pKa = 11.84GRR74 pKa = 11.84PQAVTPGRR82 pKa = 11.84QPSAGISQTRR92 pKa = 11.84SGAKK96 pKa = 9.41ISIRR100 pKa = 11.84DD101 pKa = 3.44TEE103 pKa = 4.33AFVVEE108 pKa = 4.51QGFKK112 pKa = 10.2AYY114 pKa = 9.98KK115 pKa = 9.34MNPAVNQLPRR125 pKa = 11.84LEE127 pKa = 4.34QHH129 pKa = 5.43SKK131 pKa = 8.2MYY133 pKa = 9.61KK134 pKa = 9.5RR135 pKa = 11.84YY136 pKa = 9.56RR137 pKa = 11.84IRR139 pKa = 11.84YY140 pKa = 7.83FNIAYY145 pKa = 9.77KK146 pKa = 10.35SGSGTATSGNVTVGILGGTIDD167 pKa = 4.95SKK169 pKa = 10.32VTSKK173 pKa = 11.14NVINLRR179 pKa = 11.84PSFFTPAWKK188 pKa = 10.61NEE190 pKa = 3.59TMSLGSDD197 pKa = 2.72IDD199 pKa = 3.18IARR202 pKa = 11.84WMLCEE207 pKa = 4.28QDD209 pKa = 4.85DD210 pKa = 4.04NDD212 pKa = 4.97GIAFTLYY219 pKa = 10.87VNSTEE224 pKa = 4.7AGLGMIQCSYY234 pKa = 10.58EE235 pKa = 3.93VEE237 pKa = 4.17FDD239 pKa = 3.44QPHH242 pKa = 7.15PFTT245 pKa = 4.91
MM1 pKa = 7.05MNQYY5 pKa = 10.25RR6 pKa = 11.84GRR8 pKa = 11.84GRR10 pKa = 11.84FWNAGLNLAAAAGGAAARR28 pKa = 11.84VAANRR33 pKa = 11.84FQNWVNSPPSNPYY46 pKa = 10.44NMQPIYY52 pKa = 10.27QALPPVRR59 pKa = 11.84RR60 pKa = 11.84GRR62 pKa = 11.84GRR64 pKa = 11.84GRR66 pKa = 11.84GRR68 pKa = 11.84GRR70 pKa = 11.84GRR72 pKa = 11.84GRR74 pKa = 11.84PQAVTPGRR82 pKa = 11.84QPSAGISQTRR92 pKa = 11.84SGAKK96 pKa = 9.41ISIRR100 pKa = 11.84DD101 pKa = 3.44TEE103 pKa = 4.33AFVVEE108 pKa = 4.51QGFKK112 pKa = 10.2AYY114 pKa = 9.98KK115 pKa = 9.34MNPAVNQLPRR125 pKa = 11.84LEE127 pKa = 4.34QHH129 pKa = 5.43SKK131 pKa = 8.2MYY133 pKa = 9.61KK134 pKa = 9.5RR135 pKa = 11.84YY136 pKa = 9.56RR137 pKa = 11.84IRR139 pKa = 11.84YY140 pKa = 7.83FNIAYY145 pKa = 9.77KK146 pKa = 10.35SGSGTATSGNVTVGILGGTIDD167 pKa = 4.95SKK169 pKa = 10.32VTSKK173 pKa = 11.14NVINLRR179 pKa = 11.84PSFFTPAWKK188 pKa = 10.61NEE190 pKa = 3.59TMSLGSDD197 pKa = 2.72IDD199 pKa = 3.18IARR202 pKa = 11.84WMLCEE207 pKa = 4.28QDD209 pKa = 4.85DD210 pKa = 4.04NDD212 pKa = 4.97GIAFTLYY219 pKa = 10.87VNSTEE224 pKa = 4.7AGLGMIQCSYY234 pKa = 10.58EE235 pKa = 3.93VEE237 pKa = 4.17FDD239 pKa = 3.44QPHH242 pKa = 7.15PFTT245 pKa = 4.91
Molecular weight: 27.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1502 |
215 |
558 |
375.5 |
42.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.858 ± 0.804 | 1.664 ± 0.501 |
4.927 ± 0.793 | 5.06 ± 0.654 |
3.928 ± 0.182 | 6.125 ± 0.896 |
2.796 ± 0.54 | 6.525 ± 0.548 |
5.26 ± 1.018 | 8.389 ± 1.165 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.663 ± 0.397 | 3.995 ± 0.647 |
5.792 ± 0.81 | 4.461 ± 0.411 |
7.057 ± 0.727 | 5.792 ± 0.89 |
6.059 ± 0.251 | 5.992 ± 0.331 |
2.197 ± 0.454 | 4.461 ± 0.415 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
