
Hubei lepidoptera virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Alphapaprhavirus; Hubei alphapaprhavirus
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
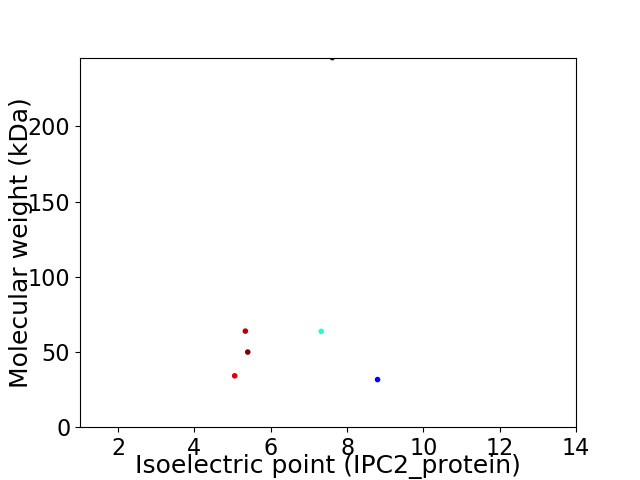
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMR9|A0A1L3KMR9_9VIRU Putative glycoprotein 1 OS=Hubei lepidoptera virus 2 OX=1922904 PE=4 SV=1
MM1 pKa = 6.71NTFRR5 pKa = 11.84RR6 pKa = 11.84INFDD10 pKa = 3.42KK11 pKa = 10.44EE12 pKa = 3.85KK13 pKa = 11.06KK14 pKa = 8.74EE15 pKa = 4.24ARR17 pKa = 11.84EE18 pKa = 3.87QNWNINLPKK27 pKa = 10.34AALEE31 pKa = 4.0QAQEE35 pKa = 4.07EE36 pKa = 4.89LGTDD40 pKa = 3.34SDD42 pKa = 4.63DD43 pKa = 3.82SAEE46 pKa = 4.16EE47 pKa = 4.02EE48 pKa = 5.04DD49 pKa = 3.91PGEE52 pKa = 5.32PKK54 pKa = 10.92DD55 pKa = 3.49PWNSVLTQNPVMSEE69 pKa = 4.09VNRR72 pKa = 11.84DD73 pKa = 3.46RR74 pKa = 11.84QFGSSDD80 pKa = 3.21IEE82 pKa = 3.96RR83 pKa = 11.84EE84 pKa = 4.28SAFSHH89 pKa = 6.75HH90 pKa = 6.38EE91 pKa = 4.25NIDD94 pKa = 3.78DD95 pKa = 3.85YY96 pKa = 11.82DD97 pKa = 4.39EE98 pKa = 4.77EE99 pKa = 4.47GHH101 pKa = 7.44AEE103 pKa = 4.24DD104 pKa = 6.15LIQSGDD110 pKa = 3.27VNQIVKK116 pKa = 10.36KK117 pKa = 10.37ALEE120 pKa = 4.26TSTPPVKK127 pKa = 10.46RR128 pKa = 11.84LGEE131 pKa = 3.89RR132 pKa = 11.84KK133 pKa = 9.33IQVNLQFMQSPPMMEE148 pKa = 3.69KK149 pKa = 9.62MKK151 pKa = 10.63DD152 pKa = 3.24IIHH155 pKa = 5.99EE156 pKa = 4.3VIKK159 pKa = 10.63AYY161 pKa = 6.79TTVPGRR167 pKa = 11.84VEE169 pKa = 4.25VSASDD174 pKa = 3.36DD175 pKa = 3.89GIYY178 pKa = 10.18IFNATNPYY186 pKa = 10.16LSPDD190 pKa = 3.62SDD192 pKa = 4.16SDD194 pKa = 3.91PLSSEE199 pKa = 3.88RR200 pKa = 11.84PYY202 pKa = 10.61PKK204 pKa = 9.86PKK206 pKa = 10.15EE207 pKa = 4.13KK208 pKa = 10.0PAPQQVQRR216 pKa = 11.84QFLPLEE222 pKa = 4.77ANTSTLTDD230 pKa = 3.39VEE232 pKa = 4.17IMIKK236 pKa = 10.3KK237 pKa = 9.73GVNFQGKK244 pKa = 9.28YY245 pKa = 9.52PGLPKK250 pKa = 10.55CIVRR254 pKa = 11.84YY255 pKa = 9.3GRR257 pKa = 11.84WGITEE262 pKa = 3.84QSVQLALKK270 pKa = 9.02EE271 pKa = 4.02NPNLSVNEE279 pKa = 4.18TIRR282 pKa = 11.84YY283 pKa = 8.94CMRR286 pKa = 11.84QAGTLKK292 pKa = 10.51KK293 pKa = 10.6FSGLYY298 pKa = 10.24KK299 pKa = 10.57FF300 pKa = 4.98
MM1 pKa = 6.71NTFRR5 pKa = 11.84RR6 pKa = 11.84INFDD10 pKa = 3.42KK11 pKa = 10.44EE12 pKa = 3.85KK13 pKa = 11.06KK14 pKa = 8.74EE15 pKa = 4.24ARR17 pKa = 11.84EE18 pKa = 3.87QNWNINLPKK27 pKa = 10.34AALEE31 pKa = 4.0QAQEE35 pKa = 4.07EE36 pKa = 4.89LGTDD40 pKa = 3.34SDD42 pKa = 4.63DD43 pKa = 3.82SAEE46 pKa = 4.16EE47 pKa = 4.02EE48 pKa = 5.04DD49 pKa = 3.91PGEE52 pKa = 5.32PKK54 pKa = 10.92DD55 pKa = 3.49PWNSVLTQNPVMSEE69 pKa = 4.09VNRR72 pKa = 11.84DD73 pKa = 3.46RR74 pKa = 11.84QFGSSDD80 pKa = 3.21IEE82 pKa = 3.96RR83 pKa = 11.84EE84 pKa = 4.28SAFSHH89 pKa = 6.75HH90 pKa = 6.38EE91 pKa = 4.25NIDD94 pKa = 3.78DD95 pKa = 3.85YY96 pKa = 11.82DD97 pKa = 4.39EE98 pKa = 4.77EE99 pKa = 4.47GHH101 pKa = 7.44AEE103 pKa = 4.24DD104 pKa = 6.15LIQSGDD110 pKa = 3.27VNQIVKK116 pKa = 10.36KK117 pKa = 10.37ALEE120 pKa = 4.26TSTPPVKK127 pKa = 10.46RR128 pKa = 11.84LGEE131 pKa = 3.89RR132 pKa = 11.84KK133 pKa = 9.33IQVNLQFMQSPPMMEE148 pKa = 3.69KK149 pKa = 9.62MKK151 pKa = 10.63DD152 pKa = 3.24IIHH155 pKa = 5.99EE156 pKa = 4.3VIKK159 pKa = 10.63AYY161 pKa = 6.79TTVPGRR167 pKa = 11.84VEE169 pKa = 4.25VSASDD174 pKa = 3.36DD175 pKa = 3.89GIYY178 pKa = 10.18IFNATNPYY186 pKa = 10.16LSPDD190 pKa = 3.62SDD192 pKa = 4.16SDD194 pKa = 3.91PLSSEE199 pKa = 3.88RR200 pKa = 11.84PYY202 pKa = 10.61PKK204 pKa = 9.86PKK206 pKa = 10.15EE207 pKa = 4.13KK208 pKa = 10.0PAPQQVQRR216 pKa = 11.84QFLPLEE222 pKa = 4.77ANTSTLTDD230 pKa = 3.39VEE232 pKa = 4.17IMIKK236 pKa = 10.3KK237 pKa = 9.73GVNFQGKK244 pKa = 9.28YY245 pKa = 9.52PGLPKK250 pKa = 10.55CIVRR254 pKa = 11.84YY255 pKa = 9.3GRR257 pKa = 11.84WGITEE262 pKa = 3.84QSVQLALKK270 pKa = 9.02EE271 pKa = 4.02NPNLSVNEE279 pKa = 4.18TIRR282 pKa = 11.84YY283 pKa = 8.94CMRR286 pKa = 11.84QAGTLKK292 pKa = 10.51KK293 pKa = 10.6FSGLYY298 pKa = 10.24KK299 pKa = 10.57FF300 pKa = 4.98
Molecular weight: 34.17 kDa
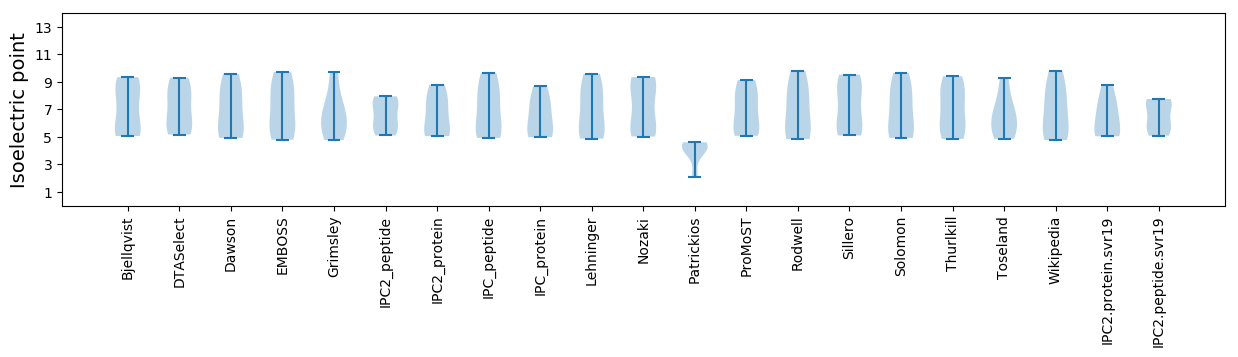
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMN7|A0A1L3KMN7_9VIRU Uncharacterized protein OS=Hubei lepidoptera virus 2 OX=1922904 PE=4 SV=1
MM1 pKa = 6.95KK2 pKa = 10.22QFVIACVKK10 pKa = 10.31PEE12 pKa = 4.02PSRR15 pKa = 11.84NSQDD19 pKa = 4.32CINSNYY25 pKa = 8.25NTTTIMLKK33 pKa = 8.91KK34 pKa = 8.62TGQGSLSSSRR44 pKa = 11.84SADD47 pKa = 3.44NEE49 pKa = 3.97IATSTDD55 pKa = 2.78SSKK58 pKa = 10.86GRR60 pKa = 11.84KK61 pKa = 8.27PKK63 pKa = 10.44NSPFSWFSKK72 pKa = 10.41KK73 pKa = 7.77PTRR76 pKa = 11.84PKK78 pKa = 9.28NHH80 pKa = 6.48QISVVKK86 pKa = 9.92PSAPPEE92 pKa = 3.93PQLRR96 pKa = 11.84FLEE99 pKa = 5.12DD100 pKa = 3.53IPVDD104 pKa = 3.18WTRR107 pKa = 11.84RR108 pKa = 11.84PEE110 pKa = 4.36DD111 pKa = 4.03EE112 pKa = 4.05SHH114 pKa = 5.93QPSIEE119 pKa = 3.9VDD121 pKa = 3.84LVSPIAFYY129 pKa = 9.8VTASLTLNSRR139 pKa = 11.84IKK141 pKa = 10.16IGSVEE146 pKa = 4.12SLLSKK151 pKa = 11.03LEE153 pKa = 3.67MWIDD157 pKa = 3.75HH158 pKa = 5.98YY159 pKa = 11.49SGSAVMYY166 pKa = 8.97PYY168 pKa = 9.57YY169 pKa = 10.15TLIYY173 pKa = 9.54ILLGSKK179 pKa = 10.35LSNRR183 pKa = 11.84PSNSKK188 pKa = 9.37MFLYY192 pKa = 10.67SSNFSGVIKK201 pKa = 10.6FGYY204 pKa = 7.95PQRR207 pKa = 11.84QGKK210 pKa = 8.11PPSEE214 pKa = 4.05TLSVLGHH221 pKa = 6.32EE222 pKa = 5.15GRR224 pKa = 11.84DD225 pKa = 3.54SNYY228 pKa = 9.9VSIRR232 pKa = 11.84FDD234 pKa = 3.07ATMKK238 pKa = 9.93EE239 pKa = 4.45SNRR242 pKa = 11.84NGVIYY247 pKa = 10.46SQLYY251 pKa = 9.25RR252 pKa = 11.84LNGYY256 pKa = 8.07NFPFPAALDD265 pKa = 3.84TFNLNIGLTHH275 pKa = 7.05EE276 pKa = 4.49GLIGPSPP283 pKa = 3.49
MM1 pKa = 6.95KK2 pKa = 10.22QFVIACVKK10 pKa = 10.31PEE12 pKa = 4.02PSRR15 pKa = 11.84NSQDD19 pKa = 4.32CINSNYY25 pKa = 8.25NTTTIMLKK33 pKa = 8.91KK34 pKa = 8.62TGQGSLSSSRR44 pKa = 11.84SADD47 pKa = 3.44NEE49 pKa = 3.97IATSTDD55 pKa = 2.78SSKK58 pKa = 10.86GRR60 pKa = 11.84KK61 pKa = 8.27PKK63 pKa = 10.44NSPFSWFSKK72 pKa = 10.41KK73 pKa = 7.77PTRR76 pKa = 11.84PKK78 pKa = 9.28NHH80 pKa = 6.48QISVVKK86 pKa = 9.92PSAPPEE92 pKa = 3.93PQLRR96 pKa = 11.84FLEE99 pKa = 5.12DD100 pKa = 3.53IPVDD104 pKa = 3.18WTRR107 pKa = 11.84RR108 pKa = 11.84PEE110 pKa = 4.36DD111 pKa = 4.03EE112 pKa = 4.05SHH114 pKa = 5.93QPSIEE119 pKa = 3.9VDD121 pKa = 3.84LVSPIAFYY129 pKa = 9.8VTASLTLNSRR139 pKa = 11.84IKK141 pKa = 10.16IGSVEE146 pKa = 4.12SLLSKK151 pKa = 11.03LEE153 pKa = 3.67MWIDD157 pKa = 3.75HH158 pKa = 5.98YY159 pKa = 11.49SGSAVMYY166 pKa = 8.97PYY168 pKa = 9.57YY169 pKa = 10.15TLIYY173 pKa = 9.54ILLGSKK179 pKa = 10.35LSNRR183 pKa = 11.84PSNSKK188 pKa = 9.37MFLYY192 pKa = 10.67SSNFSGVIKK201 pKa = 10.6FGYY204 pKa = 7.95PQRR207 pKa = 11.84QGKK210 pKa = 8.11PPSEE214 pKa = 4.05TLSVLGHH221 pKa = 6.32EE222 pKa = 5.15GRR224 pKa = 11.84DD225 pKa = 3.54SNYY228 pKa = 9.9VSIRR232 pKa = 11.84FDD234 pKa = 3.07ATMKK238 pKa = 9.93EE239 pKa = 4.45SNRR242 pKa = 11.84NGVIYY247 pKa = 10.46SQLYY251 pKa = 9.25RR252 pKa = 11.84LNGYY256 pKa = 8.07NFPFPAALDD265 pKa = 3.84TFNLNIGLTHH275 pKa = 7.05EE276 pKa = 4.49GLIGPSPP283 pKa = 3.49
Molecular weight: 31.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4293 |
283 |
2145 |
715.5 |
81.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.913 ± 0.572 | 1.77 ± 0.372 |
5.404 ± 0.266 | 6.243 ± 0.322 |
4.449 ± 0.429 | 5.777 ± 0.359 |
2.516 ± 0.192 | 7.431 ± 0.609 |
6.336 ± 0.435 | 10.04 ± 0.763 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.05 ± 0.272 | 5.334 ± 0.397 |
5.031 ± 0.487 | 3.75 ± 0.373 |
5.101 ± 0.476 | 8.642 ± 1.046 |
5.87 ± 0.273 | 5.101 ± 0.285 |
1.537 ± 0.183 | 3.704 ± 0.433 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
