
Vibrio vulnificus (strain YJ016)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio; Vibrio vulnificus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
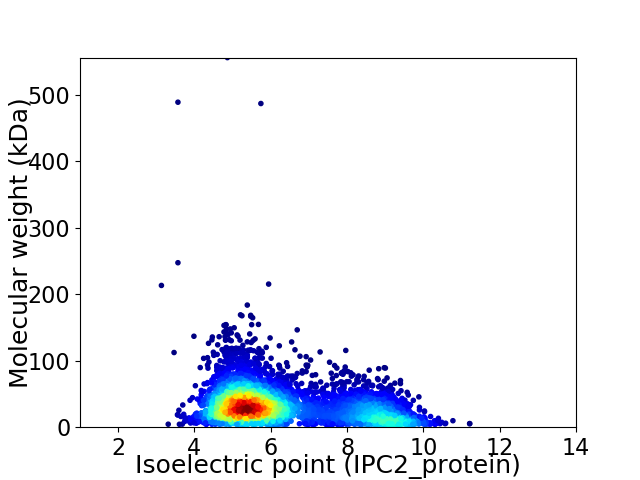
Virtual 2D-PAGE plot for 4990 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
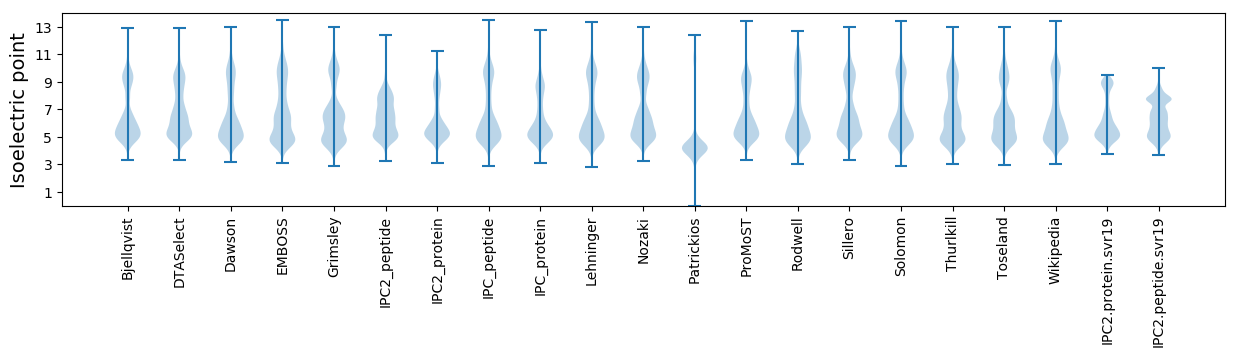
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q7MJW6|Q7MJW6_VIBVY Sodium/alanine symporter OS=Vibrio vulnificus (strain YJ016) OX=196600 GN=VV2044 PE=3 SV=1
MM1 pKa = 7.66NGLTKK6 pKa = 10.49SAISSVAAALLLAGCGGSDD25 pKa = 3.34GGKK28 pKa = 6.87TANVSFYY35 pKa = 11.37VSDD38 pKa = 4.16APVDD42 pKa = 3.92SAQSVSVGFTQVEE55 pKa = 4.24LVDD58 pKa = 3.92NEE60 pKa = 4.38GNSVFLDD67 pKa = 3.68VVPSTGVDD75 pKa = 3.47YY76 pKa = 11.36QKK78 pKa = 10.69IDD80 pKa = 3.53LLDD83 pKa = 3.69YY84 pKa = 11.06QGTDD88 pKa = 2.92SVLMVSTQPVPVGSYY103 pKa = 10.45KK104 pKa = 10.55EE105 pKa = 4.8LILHH109 pKa = 5.24VTDD112 pKa = 4.01EE113 pKa = 4.34NGVNYY118 pKa = 10.33VIDD121 pKa = 4.14EE122 pKa = 4.46NGQQPLKK129 pKa = 10.74QPSNKK134 pKa = 9.78LKK136 pKa = 10.76LGSFDD141 pKa = 3.55VTSEE145 pKa = 3.69AVQLYY150 pKa = 9.35TIEE153 pKa = 4.29FDD155 pKa = 3.6LRR157 pKa = 11.84QSLVMRR163 pKa = 11.84GNLGSNNGYY172 pKa = 9.04ILKK175 pKa = 9.28PHH177 pKa = 6.29GVSIVSNASAVSISGNVDD195 pKa = 3.41DD196 pKa = 6.03NLLTGDD202 pKa = 4.52EE203 pKa = 4.59SCTADD208 pKa = 3.27SQAFIYY214 pKa = 10.1LYY216 pKa = 10.14EE217 pKa = 5.05GEE219 pKa = 4.55GHH221 pKa = 7.36DD222 pKa = 4.25SANLIDD228 pKa = 6.13LVDD231 pKa = 4.0EE232 pKa = 5.31DD233 pKa = 5.47DD234 pKa = 5.76PEE236 pKa = 5.51FDD238 pKa = 3.52SQNPVPEE245 pKa = 4.25NAVKK249 pKa = 10.33PIASAGLDD257 pKa = 3.48EE258 pKa = 4.77NNNYY262 pKa = 10.45AFGFLNAGTYY272 pKa = 8.29TVAFSCNAGDD282 pKa = 5.1DD283 pKa = 4.14NPIQYY288 pKa = 10.48DD289 pKa = 3.65GTAIPLPTGQVGTTTLAAGEE309 pKa = 4.37NGTIDD314 pKa = 4.51FVPAPP319 pKa = 3.98
MM1 pKa = 7.66NGLTKK6 pKa = 10.49SAISSVAAALLLAGCGGSDD25 pKa = 3.34GGKK28 pKa = 6.87TANVSFYY35 pKa = 11.37VSDD38 pKa = 4.16APVDD42 pKa = 3.92SAQSVSVGFTQVEE55 pKa = 4.24LVDD58 pKa = 3.92NEE60 pKa = 4.38GNSVFLDD67 pKa = 3.68VVPSTGVDD75 pKa = 3.47YY76 pKa = 11.36QKK78 pKa = 10.69IDD80 pKa = 3.53LLDD83 pKa = 3.69YY84 pKa = 11.06QGTDD88 pKa = 2.92SVLMVSTQPVPVGSYY103 pKa = 10.45KK104 pKa = 10.55EE105 pKa = 4.8LILHH109 pKa = 5.24VTDD112 pKa = 4.01EE113 pKa = 4.34NGVNYY118 pKa = 10.33VIDD121 pKa = 4.14EE122 pKa = 4.46NGQQPLKK129 pKa = 10.74QPSNKK134 pKa = 9.78LKK136 pKa = 10.76LGSFDD141 pKa = 3.55VTSEE145 pKa = 3.69AVQLYY150 pKa = 9.35TIEE153 pKa = 4.29FDD155 pKa = 3.6LRR157 pKa = 11.84QSLVMRR163 pKa = 11.84GNLGSNNGYY172 pKa = 9.04ILKK175 pKa = 9.28PHH177 pKa = 6.29GVSIVSNASAVSISGNVDD195 pKa = 3.41DD196 pKa = 6.03NLLTGDD202 pKa = 4.52EE203 pKa = 4.59SCTADD208 pKa = 3.27SQAFIYY214 pKa = 10.1LYY216 pKa = 10.14EE217 pKa = 5.05GEE219 pKa = 4.55GHH221 pKa = 7.36DD222 pKa = 4.25SANLIDD228 pKa = 6.13LVDD231 pKa = 4.0EE232 pKa = 5.31DD233 pKa = 5.47DD234 pKa = 5.76PEE236 pKa = 5.51FDD238 pKa = 3.52SQNPVPEE245 pKa = 4.25NAVKK249 pKa = 10.33PIASAGLDD257 pKa = 3.48EE258 pKa = 4.77NNNYY262 pKa = 10.45AFGFLNAGTYY272 pKa = 8.29TVAFSCNAGDD282 pKa = 5.1DD283 pKa = 4.14NPIQYY288 pKa = 10.48DD289 pKa = 3.65GTAIPLPTGQVGTTTLAAGEE309 pKa = 4.37NGTIDD314 pKa = 4.51FVPAPP319 pKa = 3.98
Molecular weight: 33.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q7MNM8|SYI_VIBVY Isoleucine--tRNA ligase OS=Vibrio vulnificus (strain YJ016) OX=196600 GN=ileS PE=3 SV=1
MM1 pKa = 7.49LRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84VQSKK9 pKa = 10.38RR10 pKa = 11.84LNKK13 pKa = 8.79TVAANRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.13RR22 pKa = 11.84KK23 pKa = 8.96LANTKK28 pKa = 10.29RR29 pKa = 11.84KK30 pKa = 9.0ISARR34 pKa = 11.84NRR36 pKa = 11.84IFVHH40 pKa = 6.11LVNHH44 pKa = 6.98AA45 pKa = 3.49
MM1 pKa = 7.49LRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84VQSKK9 pKa = 10.38RR10 pKa = 11.84LNKK13 pKa = 8.79TVAANRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.13RR22 pKa = 11.84KK23 pKa = 8.96LANTKK28 pKa = 10.29RR29 pKa = 11.84KK30 pKa = 9.0ISARR34 pKa = 11.84NRR36 pKa = 11.84IFVHH40 pKa = 6.11LVNHH44 pKa = 6.98AA45 pKa = 3.49
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1549674 |
34 |
5206 |
310.6 |
34.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.82 ± 0.038 | 1.069 ± 0.014 |
5.269 ± 0.031 | 6.365 ± 0.041 |
4.184 ± 0.025 | 6.691 ± 0.032 |
2.363 ± 0.017 | 6.067 ± 0.028 |
5.169 ± 0.031 | 10.476 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.745 ± 0.017 | 4.117 ± 0.024 |
3.852 ± 0.025 | 4.901 ± 0.03 |
4.682 ± 0.03 | 6.551 ± 0.028 |
5.271 ± 0.031 | 7.043 ± 0.031 |
1.292 ± 0.015 | 3.074 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
