
Halomonas arcis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
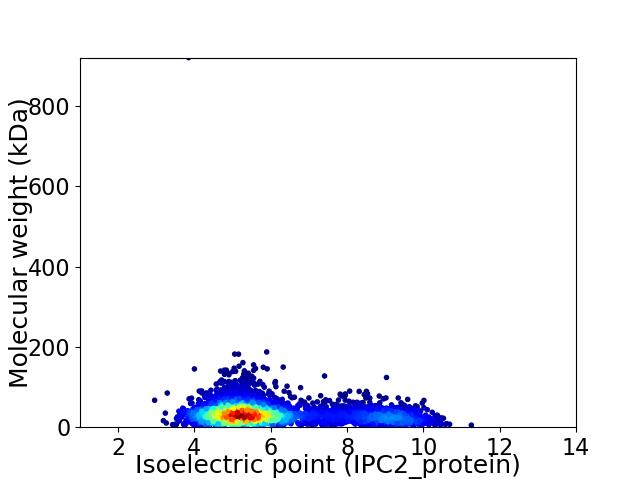
Virtual 2D-PAGE plot for 3843 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H0AQP0|A0A1H0AQP0_9GAMM Elongation factor Tu OS=Halomonas arcis OX=416873 GN=tuf PE=3 SV=1
MM1 pKa = 6.69QTLSLPRR8 pKa = 11.84TLISAAVASAFALVAVNASADD29 pKa = 3.5LSDD32 pKa = 4.96RR33 pKa = 11.84YY34 pKa = 10.77VDD36 pKa = 3.75NDD38 pKa = 3.65GDD40 pKa = 4.0LMADD44 pKa = 3.59VPSDD48 pKa = 3.26EE49 pKa = 5.06SEE51 pKa = 4.06WVDD54 pKa = 3.3PDD56 pKa = 4.01TLVFAYY62 pKa = 9.53TPVEE66 pKa = 4.43DD67 pKa = 3.79PAVYY71 pKa = 10.81ADD73 pKa = 3.38VWSDD77 pKa = 3.94FLDD80 pKa = 3.91HH81 pKa = 7.29LSDD84 pKa = 3.44ATGKK88 pKa = 9.43EE89 pKa = 4.18VQFFPVQSNAAQQEE103 pKa = 4.14ALRR106 pKa = 11.84AGRR109 pKa = 11.84LHH111 pKa = 6.08VAGFNTGGVPVAVNCGGFRR130 pKa = 11.84PFAMMAAEE138 pKa = 5.1DD139 pKa = 4.3GSFGYY144 pKa = 10.59EE145 pKa = 3.89MEE147 pKa = 5.14IITYY151 pKa = 8.53PGSGIEE157 pKa = 4.16SVEE160 pKa = 4.06DD161 pKa = 3.75LEE163 pKa = 5.04GSQLAFTSEE172 pKa = 4.03TSNSGFRR179 pKa = 11.84APSALLRR186 pKa = 11.84SEE188 pKa = 4.38YY189 pKa = 11.0GMEE192 pKa = 4.11AGEE195 pKa = 4.35DD196 pKa = 3.81FEE198 pKa = 4.97TAFSGSHH205 pKa = 6.62DD206 pKa = 3.71NSVLGVVNEE215 pKa = 4.61DD216 pKa = 3.5YY217 pKa = 10.73EE218 pKa = 4.3AAAIANSVARR228 pKa = 11.84RR229 pKa = 11.84MLARR233 pKa = 11.84GVVEE237 pKa = 4.09EE238 pKa = 4.16GDD240 pKa = 3.87YY241 pKa = 11.15EE242 pKa = 4.57VVYY245 pKa = 10.11TSQTFPTTAYY255 pKa = 10.47GLAHH259 pKa = 6.6NLDD262 pKa = 4.04PEE264 pKa = 4.32LAEE267 pKa = 5.5DD268 pKa = 3.5IQDD271 pKa = 3.89AFFSYY276 pKa = 10.36DD277 pKa = 3.06WEE279 pKa = 4.54GTALEE284 pKa = 4.51EE285 pKa = 4.03EE286 pKa = 4.67FANSGEE292 pKa = 4.1AQFIPITYY300 pKa = 8.74QEE302 pKa = 3.9NWEE305 pKa = 4.45VIRR308 pKa = 11.84TIADD312 pKa = 3.65ANGVTYY318 pKa = 10.74DD319 pKa = 3.98CDD321 pKa = 3.29
MM1 pKa = 6.69QTLSLPRR8 pKa = 11.84TLISAAVASAFALVAVNASADD29 pKa = 3.5LSDD32 pKa = 4.96RR33 pKa = 11.84YY34 pKa = 10.77VDD36 pKa = 3.75NDD38 pKa = 3.65GDD40 pKa = 4.0LMADD44 pKa = 3.59VPSDD48 pKa = 3.26EE49 pKa = 5.06SEE51 pKa = 4.06WVDD54 pKa = 3.3PDD56 pKa = 4.01TLVFAYY62 pKa = 9.53TPVEE66 pKa = 4.43DD67 pKa = 3.79PAVYY71 pKa = 10.81ADD73 pKa = 3.38VWSDD77 pKa = 3.94FLDD80 pKa = 3.91HH81 pKa = 7.29LSDD84 pKa = 3.44ATGKK88 pKa = 9.43EE89 pKa = 4.18VQFFPVQSNAAQQEE103 pKa = 4.14ALRR106 pKa = 11.84AGRR109 pKa = 11.84LHH111 pKa = 6.08VAGFNTGGVPVAVNCGGFRR130 pKa = 11.84PFAMMAAEE138 pKa = 5.1DD139 pKa = 4.3GSFGYY144 pKa = 10.59EE145 pKa = 3.89MEE147 pKa = 5.14IITYY151 pKa = 8.53PGSGIEE157 pKa = 4.16SVEE160 pKa = 4.06DD161 pKa = 3.75LEE163 pKa = 5.04GSQLAFTSEE172 pKa = 4.03TSNSGFRR179 pKa = 11.84APSALLRR186 pKa = 11.84SEE188 pKa = 4.38YY189 pKa = 11.0GMEE192 pKa = 4.11AGEE195 pKa = 4.35DD196 pKa = 3.81FEE198 pKa = 4.97TAFSGSHH205 pKa = 6.62DD206 pKa = 3.71NSVLGVVNEE215 pKa = 4.61DD216 pKa = 3.5YY217 pKa = 10.73EE218 pKa = 4.3AAAIANSVARR228 pKa = 11.84RR229 pKa = 11.84MLARR233 pKa = 11.84GVVEE237 pKa = 4.09EE238 pKa = 4.16GDD240 pKa = 3.87YY241 pKa = 11.15EE242 pKa = 4.57VVYY245 pKa = 10.11TSQTFPTTAYY255 pKa = 10.47GLAHH259 pKa = 6.6NLDD262 pKa = 4.04PEE264 pKa = 4.32LAEE267 pKa = 5.5DD268 pKa = 3.5IQDD271 pKa = 3.89AFFSYY276 pKa = 10.36DD277 pKa = 3.06WEE279 pKa = 4.54GTALEE284 pKa = 4.51EE285 pKa = 4.03EE286 pKa = 4.67FANSGEE292 pKa = 4.1AQFIPITYY300 pKa = 8.74QEE302 pKa = 3.9NWEE305 pKa = 4.45VIRR308 pKa = 11.84TIADD312 pKa = 3.65ANGVTYY318 pKa = 10.74DD319 pKa = 3.98CDD321 pKa = 3.29
Molecular weight: 34.68 kDa
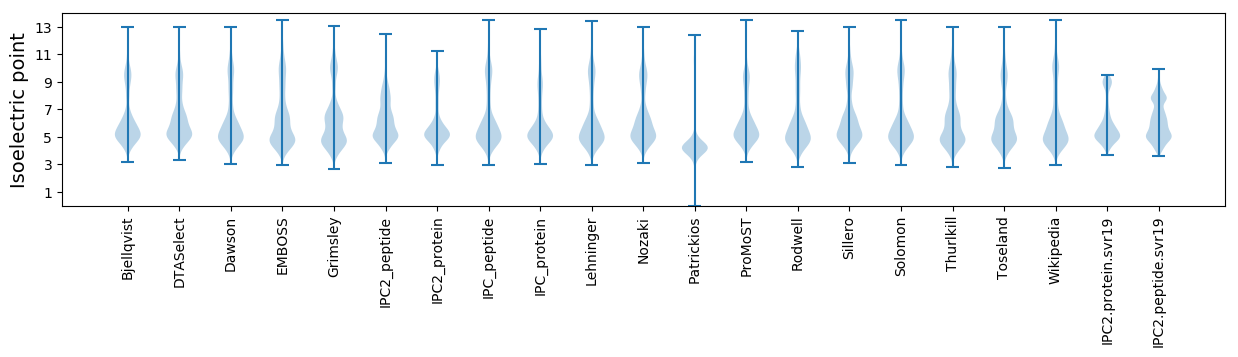
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H0IX58|A0A1H0IX58_9GAMM NAD(P)-dependent dehydrogenase short-chain alcohol dehydrogenase family OS=Halomonas arcis OX=416873 GN=SAMN04487951_12235 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.19NGRR28 pKa = 11.84AIIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.19NGRR28 pKa = 11.84AIIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1237372 |
39 |
8954 |
322.0 |
35.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.665 ± 0.041 | 0.928 ± 0.015 |
5.781 ± 0.053 | 6.188 ± 0.036 |
3.563 ± 0.028 | 7.723 ± 0.049 |
2.456 ± 0.023 | 5.056 ± 0.031 |
3.009 ± 0.031 | 11.259 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.632 ± 0.023 | 3.017 ± 0.027 |
4.751 ± 0.031 | 4.366 ± 0.035 |
6.37 ± 0.042 | 5.897 ± 0.034 |
5.244 ± 0.028 | 7.136 ± 0.031 |
1.472 ± 0.017 | 2.487 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
