
Beihai narna-like virus 23
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
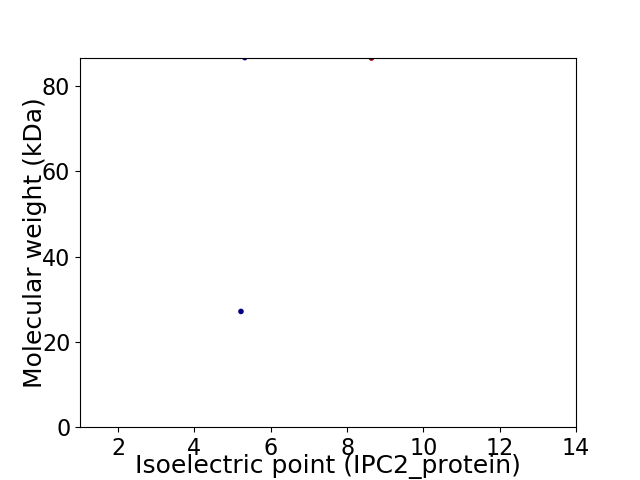
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIB7|A0A1L3KIB7_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 23 OX=1922451 PE=4 SV=1
MM1 pKa = 8.38DD2 pKa = 4.63YY3 pKa = 11.45LNMGFTFRR11 pKa = 11.84DD12 pKa = 3.66DD13 pKa = 4.03RR14 pKa = 11.84SIPDD18 pKa = 3.97DD19 pKa = 4.09PNEE22 pKa = 3.96GFTRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84FIEE31 pKa = 4.1EE32 pKa = 4.22LVLSIQTGSPPPAALEE48 pKa = 4.72DD49 pKa = 3.88GLHH52 pKa = 6.16RR53 pKa = 11.84FVEE56 pKa = 4.32ADD58 pKa = 3.19SYY60 pKa = 11.42LLHH63 pKa = 6.44VLSKK67 pKa = 11.05SQDD70 pKa = 3.31LPNILFLVSRR80 pKa = 11.84DD81 pKa = 3.1KK82 pKa = 11.18RR83 pKa = 11.84LALRR87 pKa = 11.84MRR89 pKa = 11.84DD90 pKa = 3.4VARR93 pKa = 11.84GVVTRR98 pKa = 11.84EE99 pKa = 3.21RR100 pKa = 11.84GYY102 pKa = 9.8NASRR106 pKa = 11.84DD107 pKa = 3.71VVIYY111 pKa = 9.79MVDD114 pKa = 3.24PADD117 pKa = 3.87FRR119 pKa = 11.84FGYY122 pKa = 9.14MNEE125 pKa = 5.05HH126 pKa = 6.29IAHH129 pKa = 5.88WTGGYY134 pKa = 9.17PDD136 pKa = 3.84SKK138 pKa = 9.97VPYY141 pKa = 9.91RR142 pKa = 11.84EE143 pKa = 3.97FVDD146 pKa = 4.7FGQVNFLAFTKK157 pKa = 10.72DD158 pKa = 3.63PIDD161 pKa = 3.77PFYY164 pKa = 9.96EE165 pKa = 4.05TEE167 pKa = 4.67RR168 pKa = 11.84IHH170 pKa = 7.28AVPCKK175 pKa = 10.08IRR177 pKa = 11.84GDD179 pKa = 3.88LVPGVYY185 pKa = 9.94HH186 pKa = 6.06SEE188 pKa = 4.18RR189 pKa = 11.84VCADD193 pKa = 3.61YY194 pKa = 11.28QDD196 pKa = 3.76TEE198 pKa = 4.65YY199 pKa = 10.68PFLSLTTQPLEE210 pKa = 4.12TILRR214 pKa = 11.84DD215 pKa = 3.55GVRR218 pKa = 11.84YY219 pKa = 8.11PHH221 pKa = 6.84IPSFARR227 pKa = 11.84DD228 pKa = 3.48LSAPTTWW235 pKa = 3.26
MM1 pKa = 8.38DD2 pKa = 4.63YY3 pKa = 11.45LNMGFTFRR11 pKa = 11.84DD12 pKa = 3.66DD13 pKa = 4.03RR14 pKa = 11.84SIPDD18 pKa = 3.97DD19 pKa = 4.09PNEE22 pKa = 3.96GFTRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84FIEE31 pKa = 4.1EE32 pKa = 4.22LVLSIQTGSPPPAALEE48 pKa = 4.72DD49 pKa = 3.88GLHH52 pKa = 6.16RR53 pKa = 11.84FVEE56 pKa = 4.32ADD58 pKa = 3.19SYY60 pKa = 11.42LLHH63 pKa = 6.44VLSKK67 pKa = 11.05SQDD70 pKa = 3.31LPNILFLVSRR80 pKa = 11.84DD81 pKa = 3.1KK82 pKa = 11.18RR83 pKa = 11.84LALRR87 pKa = 11.84MRR89 pKa = 11.84DD90 pKa = 3.4VARR93 pKa = 11.84GVVTRR98 pKa = 11.84EE99 pKa = 3.21RR100 pKa = 11.84GYY102 pKa = 9.8NASRR106 pKa = 11.84DD107 pKa = 3.71VVIYY111 pKa = 9.79MVDD114 pKa = 3.24PADD117 pKa = 3.87FRR119 pKa = 11.84FGYY122 pKa = 9.14MNEE125 pKa = 5.05HH126 pKa = 6.29IAHH129 pKa = 5.88WTGGYY134 pKa = 9.17PDD136 pKa = 3.84SKK138 pKa = 9.97VPYY141 pKa = 9.91RR142 pKa = 11.84EE143 pKa = 3.97FVDD146 pKa = 4.7FGQVNFLAFTKK157 pKa = 10.72DD158 pKa = 3.63PIDD161 pKa = 3.77PFYY164 pKa = 9.96EE165 pKa = 4.05TEE167 pKa = 4.67RR168 pKa = 11.84IHH170 pKa = 7.28AVPCKK175 pKa = 10.08IRR177 pKa = 11.84GDD179 pKa = 3.88LVPGVYY185 pKa = 9.94HH186 pKa = 6.06SEE188 pKa = 4.18RR189 pKa = 11.84VCADD193 pKa = 3.61YY194 pKa = 11.28QDD196 pKa = 3.76TEE198 pKa = 4.65YY199 pKa = 10.68PFLSLTTQPLEE210 pKa = 4.12TILRR214 pKa = 11.84DD215 pKa = 3.55GVRR218 pKa = 11.84YY219 pKa = 8.11PHH221 pKa = 6.84IPSFARR227 pKa = 11.84DD228 pKa = 3.48LSAPTTWW235 pKa = 3.26
Molecular weight: 27.14 kDa
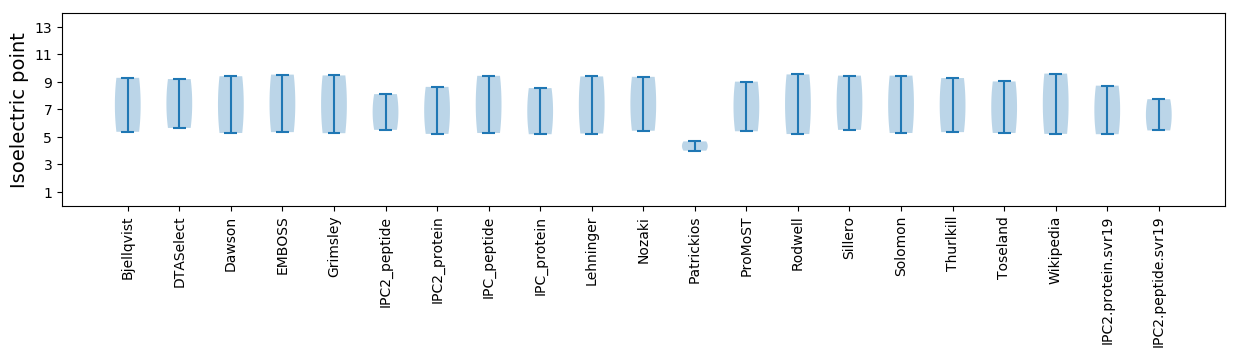
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIB7|A0A1L3KIB7_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 23 OX=1922451 PE=4 SV=1
MM1 pKa = 7.48SGHH4 pKa = 6.01FQRR7 pKa = 11.84ALSKK11 pKa = 10.61LCRR14 pKa = 11.84SYY16 pKa = 11.33DD17 pKa = 3.32YY18 pKa = 10.37RR19 pKa = 11.84TIQRR23 pKa = 11.84WWYY26 pKa = 7.85TVDD29 pKa = 4.18GLVLPLLLTHH39 pKa = 7.31PDD41 pKa = 3.68PDD43 pKa = 3.92SQMEE47 pKa = 4.46TIDD50 pKa = 3.83RR51 pKa = 11.84LIKK54 pKa = 10.61FGFEE58 pKa = 3.66NCANNYY64 pKa = 7.59SHH66 pKa = 7.87FISRR70 pKa = 11.84MKK72 pKa = 10.4RR73 pKa = 11.84LKK75 pKa = 10.47KK76 pKa = 10.34LLRR79 pKa = 11.84KK80 pKa = 9.34EE81 pKa = 4.05FAEE84 pKa = 4.61GTDD87 pKa = 3.67LTSGQLFVDD96 pKa = 4.34LSTYY100 pKa = 10.42RR101 pKa = 11.84KK102 pKa = 9.48VFYY105 pKa = 10.27RR106 pKa = 11.84YY107 pKa = 10.14LDD109 pKa = 3.44LTLSEE114 pKa = 4.84RR115 pKa = 11.84SATEE119 pKa = 4.37CVMAWTQARR128 pKa = 11.84AAGLADD134 pKa = 3.09RR135 pKa = 11.84RR136 pKa = 11.84MIEE139 pKa = 3.8KK140 pKa = 10.38SLEE143 pKa = 4.08KK144 pKa = 10.74AEE146 pKa = 4.57KK147 pKa = 9.39TLSTPRR153 pKa = 11.84RR154 pKa = 11.84VYY156 pKa = 10.76LRR158 pKa = 11.84GRR160 pKa = 11.84WRR162 pKa = 11.84EE163 pKa = 3.58IHH165 pKa = 6.2AHH167 pKa = 4.56YY168 pKa = 10.5QGFIPVLEE176 pKa = 4.41PEE178 pKa = 4.63VPRR181 pKa = 11.84GPLLDD186 pKa = 3.5VVRR189 pKa = 11.84RR190 pKa = 11.84VANKK194 pKa = 9.25VRR196 pKa = 11.84GEE198 pKa = 4.07TAHH201 pKa = 7.01ISTGPHH207 pKa = 5.54ACLEE211 pKa = 4.16IKK213 pKa = 10.24QAFGGQAAGIAYY225 pKa = 9.67LCKK228 pKa = 9.88RR229 pKa = 11.84DD230 pKa = 3.93VLTVSYY236 pKa = 11.09DD237 pKa = 3.34PVTLEE242 pKa = 4.15RR243 pKa = 11.84TFHH246 pKa = 6.83PARR249 pKa = 11.84PCASAQDD256 pKa = 4.18LLSWAISRR264 pKa = 11.84ALDD267 pKa = 3.74APVHH271 pKa = 4.83TRR273 pKa = 11.84MVRR276 pKa = 11.84IVAVSEE282 pKa = 3.78PSKK285 pKa = 11.12ARR287 pKa = 11.84TVTVGALAYY296 pKa = 9.71QIILGVVSKK305 pKa = 10.67IFQPALASGISRR317 pKa = 11.84TGLEE321 pKa = 4.24GTRR324 pKa = 11.84NLYY327 pKa = 9.41EE328 pKa = 4.2ALNNDD333 pKa = 4.03FDD335 pKa = 4.78PSNGLWGPLRR345 pKa = 11.84DD346 pKa = 3.6MDD348 pKa = 3.85RR349 pKa = 11.84TGKK352 pKa = 10.4FPVFALTSDD361 pKa = 3.77LEE363 pKa = 4.18EE364 pKa = 4.16ATDD367 pKa = 3.84YY368 pKa = 11.88GDD370 pKa = 3.92LAVARR375 pKa = 11.84QILQALLMRR384 pKa = 11.84CRR386 pKa = 11.84DD387 pKa = 3.3IPGFPLGLAVLAKK400 pKa = 10.46SLFLSSRR407 pKa = 11.84IIIRR411 pKa = 11.84PGYY414 pKa = 9.77GHH416 pKa = 7.1GLVHH420 pKa = 7.4WFRR423 pKa = 11.84KK424 pKa = 8.87RR425 pKa = 11.84NGWLMGDD432 pKa = 3.53RR433 pKa = 11.84MTKK436 pKa = 9.96VVLTLAHH443 pKa = 6.89EE444 pKa = 4.3IGILSAGIQFARR456 pKa = 11.84ICGDD460 pKa = 3.55DD461 pKa = 3.6VFALSKK467 pKa = 11.05APAQLQRR474 pKa = 11.84YY475 pKa = 7.84HH476 pKa = 8.26KK477 pKa = 11.08VMTDD481 pKa = 3.22LGFKK485 pKa = 10.19ISEE488 pKa = 4.36DD489 pKa = 3.16DD490 pKa = 3.52HH491 pKa = 6.8FISRR495 pKa = 11.84RR496 pKa = 11.84ILFYY500 pKa = 10.92CEE502 pKa = 3.82EE503 pKa = 4.22VSLVPQDD510 pKa = 4.84AKK512 pKa = 11.16DD513 pKa = 3.76LPSVCNRR520 pKa = 11.84RR521 pKa = 11.84SEE523 pKa = 4.03PSCYY527 pKa = 9.88VDD529 pKa = 3.3YY530 pKa = 10.51PRR532 pKa = 11.84IRR534 pKa = 11.84LLLPIKK540 pKa = 10.82VEE542 pKa = 4.36TNALSYY548 pKa = 10.71TDD550 pKa = 3.04TGRR553 pKa = 11.84FHH555 pKa = 7.44LLGKK559 pKa = 8.41EE560 pKa = 3.97MRR562 pKa = 11.84WVFQNSSQQADD573 pKa = 3.62PFIRR577 pKa = 11.84ASLLQHH583 pKa = 6.71IAIAMPRR590 pKa = 11.84DD591 pKa = 3.44ILSPFLPQEE600 pKa = 3.98LAGDD604 pKa = 3.87GAFPHH609 pKa = 6.31SAEE612 pKa = 4.23FLLSVIEE619 pKa = 4.31RR620 pKa = 11.84KK621 pKa = 10.11SINYY625 pKa = 9.18DD626 pKa = 2.97EE627 pKa = 4.49CLYY630 pKa = 10.43RR631 pKa = 11.84IHH633 pKa = 7.09SLTHH637 pKa = 5.68GKK639 pKa = 8.71WGFRR643 pKa = 11.84YY644 pKa = 9.76LRR646 pKa = 11.84ADD648 pKa = 3.87NINEE652 pKa = 4.18VVHH655 pKa = 6.93KK656 pKa = 7.97YY657 pKa = 10.17HH658 pKa = 7.4QMVPKK663 pKa = 10.47LKK665 pKa = 10.31VLEE668 pKa = 4.46SVLPPDD674 pKa = 3.51AVVRR678 pKa = 11.84ASEE681 pKa = 4.1VLISSLKK688 pKa = 10.17VKK690 pKa = 10.49GLEE693 pKa = 4.25TPEE696 pKa = 3.76KK697 pKa = 10.09TFFRR701 pKa = 11.84LYY703 pKa = 10.43RR704 pKa = 11.84SYY706 pKa = 10.2YY707 pKa = 7.36WYY709 pKa = 10.65KK710 pKa = 10.26VLHH713 pKa = 5.91GMKK716 pKa = 10.43APVLNFDD723 pKa = 3.53EE724 pKa = 5.19DD725 pKa = 3.46RR726 pKa = 11.84TRR728 pKa = 11.84VHH730 pKa = 7.05GGRR733 pKa = 11.84EE734 pKa = 4.11FKK736 pKa = 10.92LDD738 pKa = 3.52IPPDD742 pKa = 3.54VLVEE746 pKa = 4.1RR747 pKa = 11.84FYY749 pKa = 9.29TTWRR753 pKa = 11.84DD754 pKa = 3.23SGFTFF759 pKa = 4.98
MM1 pKa = 7.48SGHH4 pKa = 6.01FQRR7 pKa = 11.84ALSKK11 pKa = 10.61LCRR14 pKa = 11.84SYY16 pKa = 11.33DD17 pKa = 3.32YY18 pKa = 10.37RR19 pKa = 11.84TIQRR23 pKa = 11.84WWYY26 pKa = 7.85TVDD29 pKa = 4.18GLVLPLLLTHH39 pKa = 7.31PDD41 pKa = 3.68PDD43 pKa = 3.92SQMEE47 pKa = 4.46TIDD50 pKa = 3.83RR51 pKa = 11.84LIKK54 pKa = 10.61FGFEE58 pKa = 3.66NCANNYY64 pKa = 7.59SHH66 pKa = 7.87FISRR70 pKa = 11.84MKK72 pKa = 10.4RR73 pKa = 11.84LKK75 pKa = 10.47KK76 pKa = 10.34LLRR79 pKa = 11.84KK80 pKa = 9.34EE81 pKa = 4.05FAEE84 pKa = 4.61GTDD87 pKa = 3.67LTSGQLFVDD96 pKa = 4.34LSTYY100 pKa = 10.42RR101 pKa = 11.84KK102 pKa = 9.48VFYY105 pKa = 10.27RR106 pKa = 11.84YY107 pKa = 10.14LDD109 pKa = 3.44LTLSEE114 pKa = 4.84RR115 pKa = 11.84SATEE119 pKa = 4.37CVMAWTQARR128 pKa = 11.84AAGLADD134 pKa = 3.09RR135 pKa = 11.84RR136 pKa = 11.84MIEE139 pKa = 3.8KK140 pKa = 10.38SLEE143 pKa = 4.08KK144 pKa = 10.74AEE146 pKa = 4.57KK147 pKa = 9.39TLSTPRR153 pKa = 11.84RR154 pKa = 11.84VYY156 pKa = 10.76LRR158 pKa = 11.84GRR160 pKa = 11.84WRR162 pKa = 11.84EE163 pKa = 3.58IHH165 pKa = 6.2AHH167 pKa = 4.56YY168 pKa = 10.5QGFIPVLEE176 pKa = 4.41PEE178 pKa = 4.63VPRR181 pKa = 11.84GPLLDD186 pKa = 3.5VVRR189 pKa = 11.84RR190 pKa = 11.84VANKK194 pKa = 9.25VRR196 pKa = 11.84GEE198 pKa = 4.07TAHH201 pKa = 7.01ISTGPHH207 pKa = 5.54ACLEE211 pKa = 4.16IKK213 pKa = 10.24QAFGGQAAGIAYY225 pKa = 9.67LCKK228 pKa = 9.88RR229 pKa = 11.84DD230 pKa = 3.93VLTVSYY236 pKa = 11.09DD237 pKa = 3.34PVTLEE242 pKa = 4.15RR243 pKa = 11.84TFHH246 pKa = 6.83PARR249 pKa = 11.84PCASAQDD256 pKa = 4.18LLSWAISRR264 pKa = 11.84ALDD267 pKa = 3.74APVHH271 pKa = 4.83TRR273 pKa = 11.84MVRR276 pKa = 11.84IVAVSEE282 pKa = 3.78PSKK285 pKa = 11.12ARR287 pKa = 11.84TVTVGALAYY296 pKa = 9.71QIILGVVSKK305 pKa = 10.67IFQPALASGISRR317 pKa = 11.84TGLEE321 pKa = 4.24GTRR324 pKa = 11.84NLYY327 pKa = 9.41EE328 pKa = 4.2ALNNDD333 pKa = 4.03FDD335 pKa = 4.78PSNGLWGPLRR345 pKa = 11.84DD346 pKa = 3.6MDD348 pKa = 3.85RR349 pKa = 11.84TGKK352 pKa = 10.4FPVFALTSDD361 pKa = 3.77LEE363 pKa = 4.18EE364 pKa = 4.16ATDD367 pKa = 3.84YY368 pKa = 11.88GDD370 pKa = 3.92LAVARR375 pKa = 11.84QILQALLMRR384 pKa = 11.84CRR386 pKa = 11.84DD387 pKa = 3.3IPGFPLGLAVLAKK400 pKa = 10.46SLFLSSRR407 pKa = 11.84IIIRR411 pKa = 11.84PGYY414 pKa = 9.77GHH416 pKa = 7.1GLVHH420 pKa = 7.4WFRR423 pKa = 11.84KK424 pKa = 8.87RR425 pKa = 11.84NGWLMGDD432 pKa = 3.53RR433 pKa = 11.84MTKK436 pKa = 9.96VVLTLAHH443 pKa = 6.89EE444 pKa = 4.3IGILSAGIQFARR456 pKa = 11.84ICGDD460 pKa = 3.55DD461 pKa = 3.6VFALSKK467 pKa = 11.05APAQLQRR474 pKa = 11.84YY475 pKa = 7.84HH476 pKa = 8.26KK477 pKa = 11.08VMTDD481 pKa = 3.22LGFKK485 pKa = 10.19ISEE488 pKa = 4.36DD489 pKa = 3.16DD490 pKa = 3.52HH491 pKa = 6.8FISRR495 pKa = 11.84RR496 pKa = 11.84ILFYY500 pKa = 10.92CEE502 pKa = 3.82EE503 pKa = 4.22VSLVPQDD510 pKa = 4.84AKK512 pKa = 11.16DD513 pKa = 3.76LPSVCNRR520 pKa = 11.84RR521 pKa = 11.84SEE523 pKa = 4.03PSCYY527 pKa = 9.88VDD529 pKa = 3.3YY530 pKa = 10.51PRR532 pKa = 11.84IRR534 pKa = 11.84LLLPIKK540 pKa = 10.82VEE542 pKa = 4.36TNALSYY548 pKa = 10.71TDD550 pKa = 3.04TGRR553 pKa = 11.84FHH555 pKa = 7.44LLGKK559 pKa = 8.41EE560 pKa = 3.97MRR562 pKa = 11.84WVFQNSSQQADD573 pKa = 3.62PFIRR577 pKa = 11.84ASLLQHH583 pKa = 6.71IAIAMPRR590 pKa = 11.84DD591 pKa = 3.44ILSPFLPQEE600 pKa = 3.98LAGDD604 pKa = 3.87GAFPHH609 pKa = 6.31SAEE612 pKa = 4.23FLLSVIEE619 pKa = 4.31RR620 pKa = 11.84KK621 pKa = 10.11SINYY625 pKa = 9.18DD626 pKa = 2.97EE627 pKa = 4.49CLYY630 pKa = 10.43RR631 pKa = 11.84IHH633 pKa = 7.09SLTHH637 pKa = 5.68GKK639 pKa = 8.71WGFRR643 pKa = 11.84YY644 pKa = 9.76LRR646 pKa = 11.84ADD648 pKa = 3.87NINEE652 pKa = 4.18VVHH655 pKa = 6.93KK656 pKa = 7.97YY657 pKa = 10.17HH658 pKa = 7.4QMVPKK663 pKa = 10.47LKK665 pKa = 10.31VLEE668 pKa = 4.46SVLPPDD674 pKa = 3.51AVVRR678 pKa = 11.84ASEE681 pKa = 4.1VLISSLKK688 pKa = 10.17VKK690 pKa = 10.49GLEE693 pKa = 4.25TPEE696 pKa = 3.76KK697 pKa = 10.09TFFRR701 pKa = 11.84LYY703 pKa = 10.43RR704 pKa = 11.84SYY706 pKa = 10.2YY707 pKa = 7.36WYY709 pKa = 10.65KK710 pKa = 10.26VLHH713 pKa = 5.91GMKK716 pKa = 10.43APVLNFDD723 pKa = 3.53EE724 pKa = 5.19DD725 pKa = 3.46RR726 pKa = 11.84TRR728 pKa = 11.84VHH730 pKa = 7.05GGRR733 pKa = 11.84EE734 pKa = 4.11FKK736 pKa = 10.92LDD738 pKa = 3.52IPPDD742 pKa = 3.54VLVEE746 pKa = 4.1RR747 pKa = 11.84FYY749 pKa = 9.29TTWRR753 pKa = 11.84DD754 pKa = 3.23SGFTFF759 pKa = 4.98
Molecular weight: 86.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
994 |
235 |
759 |
497.0 |
56.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.042 ± 0.736 | 1.408 ± 0.272 |
6.539 ± 1.376 | 5.231 ± 0.146 |
5.131 ± 0.61 | 5.835 ± 0.148 |
3.018 ± 0.019 | 5.131 ± 0.219 |
4.125 ± 0.973 | 10.865 ± 1.148 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.012 ± 0.056 | 2.213 ± 0.166 |
5.734 ± 0.938 | 2.817 ± 0.336 |
8.853 ± 0.248 | 6.338 ± 0.393 |
5.131 ± 0.195 | 7.042 ± 0.301 |
1.408 ± 0.272 | 4.125 ± 0.478 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
