
Nostoc sphaeroides CCNUC1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Nostoc; Nostoc sphaeroides
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
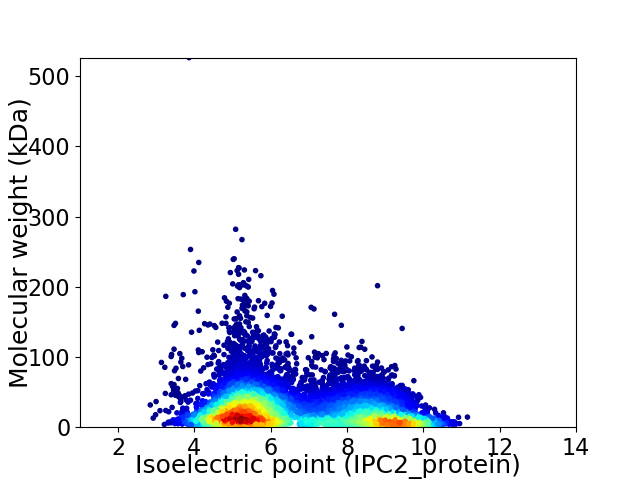
Virtual 2D-PAGE plot for 10233 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
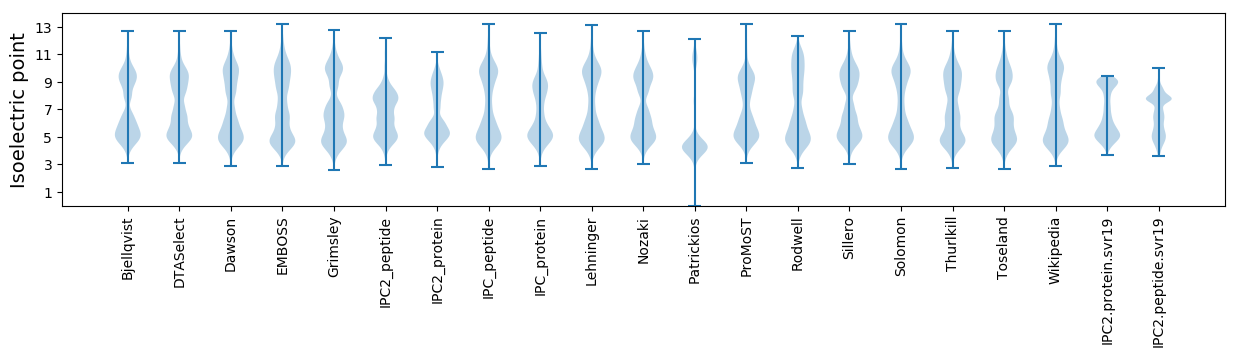
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5P8W931|A0A5P8W931_9NOSO 3-methyl-2-oxobutanoate hydroxymethyltransferase OS=Nostoc sphaeroides CCNUC1 OX=2653204 GN=panB PE=3 SV=1
MM1 pKa = 7.99DD2 pKa = 3.72MALIKK7 pKa = 10.82GSLGNDD13 pKa = 3.06NLTGTPEE20 pKa = 4.22ADD22 pKa = 4.32SIFGDD27 pKa = 4.01LGDD30 pKa = 6.2DD31 pKa = 3.52ILDD34 pKa = 4.06GLGGDD39 pKa = 4.65DD40 pKa = 4.69KK41 pKa = 11.5FVFNNVIDD49 pKa = 3.91VLVFDD54 pKa = 5.55DD55 pKa = 6.33FSDD58 pKa = 4.21LYY60 pKa = 10.58IYY62 pKa = 9.87STDD65 pKa = 3.16GFDD68 pKa = 3.52IIKK71 pKa = 10.33NFFSGEE77 pKa = 4.23VISIDD82 pKa = 3.44PVNHH86 pKa = 5.59NRR88 pKa = 11.84KK89 pKa = 8.94HH90 pKa = 6.72VDD92 pKa = 2.75INVVISAAPAVVFPSEE108 pKa = 4.24KK109 pKa = 9.84TEE111 pKa = 4.31SIYY114 pKa = 10.52GTTGSDD120 pKa = 3.72TIQGTSADD128 pKa = 3.98EE129 pKa = 4.27IIYY132 pKa = 10.32GLEE135 pKa = 4.35GNDD138 pKa = 3.18VLYY141 pKa = 11.31GNDD144 pKa = 4.83GYY146 pKa = 11.76DD147 pKa = 3.23MLYY150 pKa = 10.67GGSGNDD156 pKa = 3.36SLYY159 pKa = 11.29GGANFGDD166 pKa = 4.22EE167 pKa = 4.7LYY169 pKa = 11.11GGLGNDD175 pKa = 3.65YY176 pKa = 11.09LNGGGANVLYY186 pKa = 10.73GEE188 pKa = 4.73EE189 pKa = 4.62GDD191 pKa = 5.79DD192 pKa = 4.19IFDD195 pKa = 5.2GIDD198 pKa = 4.07GIDD201 pKa = 4.48GIDD204 pKa = 4.3GIDD207 pKa = 3.54GVKK210 pKa = 10.12VHH212 pKa = 6.93IIEE215 pKa = 4.38YY216 pKa = 9.93LNQGTDD222 pKa = 3.31TVQASSSYY230 pKa = 11.09NLGDD234 pKa = 3.48NLEE237 pKa = 4.13NLVLTGSSDD246 pKa = 2.97IDD248 pKa = 4.29GIGNALDD255 pKa = 3.71NRR257 pKa = 11.84IEE259 pKa = 4.26GNAGHH264 pKa = 6.63NFLIGGDD271 pKa = 3.82GNDD274 pKa = 3.45YY275 pKa = 11.11LLGYY279 pKa = 9.98GDD281 pKa = 4.37YY282 pKa = 10.83DD283 pKa = 3.59ILVGEE288 pKa = 4.5AGKK291 pKa = 8.9DD292 pKa = 3.51TLVGGAGNDD301 pKa = 3.66FLNGGSGNDD310 pKa = 3.83FLNGGAGNNTLTGDD324 pKa = 3.53AGADD328 pKa = 2.94TFVFNFRR335 pKa = 11.84SEE337 pKa = 4.44GIDD340 pKa = 3.14IIKK343 pKa = 10.19DD344 pKa = 3.38FRR346 pKa = 11.84YY347 pKa = 9.77LQSDD351 pKa = 4.55KK352 pKa = 11.05IQISTIGFGATSTKK366 pKa = 9.93EE367 pKa = 3.42FSYY370 pKa = 11.11NRR372 pKa = 11.84NTGALSFQGTQFATLEE388 pKa = 4.17NKK390 pKa = 10.09PNFIPRR396 pKa = 11.84LDD398 pKa = 3.64IEE400 pKa = 4.89LVSEE404 pKa = 4.71DD405 pKa = 3.88LSSSISYY412 pKa = 10.41ILL414 pKa = 3.82
MM1 pKa = 7.99DD2 pKa = 3.72MALIKK7 pKa = 10.82GSLGNDD13 pKa = 3.06NLTGTPEE20 pKa = 4.22ADD22 pKa = 4.32SIFGDD27 pKa = 4.01LGDD30 pKa = 6.2DD31 pKa = 3.52ILDD34 pKa = 4.06GLGGDD39 pKa = 4.65DD40 pKa = 4.69KK41 pKa = 11.5FVFNNVIDD49 pKa = 3.91VLVFDD54 pKa = 5.55DD55 pKa = 6.33FSDD58 pKa = 4.21LYY60 pKa = 10.58IYY62 pKa = 9.87STDD65 pKa = 3.16GFDD68 pKa = 3.52IIKK71 pKa = 10.33NFFSGEE77 pKa = 4.23VISIDD82 pKa = 3.44PVNHH86 pKa = 5.59NRR88 pKa = 11.84KK89 pKa = 8.94HH90 pKa = 6.72VDD92 pKa = 2.75INVVISAAPAVVFPSEE108 pKa = 4.24KK109 pKa = 9.84TEE111 pKa = 4.31SIYY114 pKa = 10.52GTTGSDD120 pKa = 3.72TIQGTSADD128 pKa = 3.98EE129 pKa = 4.27IIYY132 pKa = 10.32GLEE135 pKa = 4.35GNDD138 pKa = 3.18VLYY141 pKa = 11.31GNDD144 pKa = 4.83GYY146 pKa = 11.76DD147 pKa = 3.23MLYY150 pKa = 10.67GGSGNDD156 pKa = 3.36SLYY159 pKa = 11.29GGANFGDD166 pKa = 4.22EE167 pKa = 4.7LYY169 pKa = 11.11GGLGNDD175 pKa = 3.65YY176 pKa = 11.09LNGGGANVLYY186 pKa = 10.73GEE188 pKa = 4.73EE189 pKa = 4.62GDD191 pKa = 5.79DD192 pKa = 4.19IFDD195 pKa = 5.2GIDD198 pKa = 4.07GIDD201 pKa = 4.48GIDD204 pKa = 4.3GIDD207 pKa = 3.54GVKK210 pKa = 10.12VHH212 pKa = 6.93IIEE215 pKa = 4.38YY216 pKa = 9.93LNQGTDD222 pKa = 3.31TVQASSSYY230 pKa = 11.09NLGDD234 pKa = 3.48NLEE237 pKa = 4.13NLVLTGSSDD246 pKa = 2.97IDD248 pKa = 4.29GIGNALDD255 pKa = 3.71NRR257 pKa = 11.84IEE259 pKa = 4.26GNAGHH264 pKa = 6.63NFLIGGDD271 pKa = 3.82GNDD274 pKa = 3.45YY275 pKa = 11.11LLGYY279 pKa = 9.98GDD281 pKa = 4.37YY282 pKa = 10.83DD283 pKa = 3.59ILVGEE288 pKa = 4.5AGKK291 pKa = 8.9DD292 pKa = 3.51TLVGGAGNDD301 pKa = 3.66FLNGGSGNDD310 pKa = 3.83FLNGGAGNNTLTGDD324 pKa = 3.53AGADD328 pKa = 2.94TFVFNFRR335 pKa = 11.84SEE337 pKa = 4.44GIDD340 pKa = 3.14IIKK343 pKa = 10.19DD344 pKa = 3.38FRR346 pKa = 11.84YY347 pKa = 9.77LQSDD351 pKa = 4.55KK352 pKa = 11.05IQISTIGFGATSTKK366 pKa = 9.93EE367 pKa = 3.42FSYY370 pKa = 11.11NRR372 pKa = 11.84NTGALSFQGTQFATLEE388 pKa = 4.17NKK390 pKa = 10.09PNFIPRR396 pKa = 11.84LDD398 pKa = 3.64IEE400 pKa = 4.89LVSEE404 pKa = 4.71DD405 pKa = 3.88LSSSISYY412 pKa = 10.41ILL414 pKa = 3.82
Molecular weight: 43.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5P8W2P0|A0A5P8W2P0_9NOSO DNA-processing protein DprA OS=Nostoc sphaeroides CCNUC1 OX=2653204 GN=GXM_04444 PE=3 SV=1
MM1 pKa = 7.66LFRR4 pKa = 11.84RR5 pKa = 11.84THH7 pKa = 5.36LLFRR11 pKa = 11.84RR12 pKa = 11.84THH14 pKa = 5.49LLFRR18 pKa = 11.84RR19 pKa = 11.84THH21 pKa = 5.49LLFRR25 pKa = 11.84RR26 pKa = 11.84THH28 pKa = 5.49LLFRR32 pKa = 11.84RR33 pKa = 11.84THH35 pKa = 5.49LLFRR39 pKa = 11.84RR40 pKa = 11.84THH42 pKa = 5.49LLFRR46 pKa = 11.84RR47 pKa = 11.84THH49 pKa = 5.49LLFRR53 pKa = 11.84RR54 pKa = 11.84THH56 pKa = 5.49LLFRR60 pKa = 11.84RR61 pKa = 11.84THH63 pKa = 5.49LLFRR67 pKa = 11.84RR68 pKa = 11.84THH70 pKa = 5.36LLFRR74 pKa = 11.84RR75 pKa = 11.84VHH77 pKa = 5.46NAPYY81 pKa = 10.16KK82 pKa = 7.48ITRR85 pKa = 11.84RR86 pKa = 11.84VEE88 pKa = 3.95TYY90 pKa = 8.97TGHH93 pKa = 6.0NTVQLSPKK101 pKa = 9.33TLVKK105 pKa = 9.75TRR107 pKa = 11.84NFASLQVV114 pKa = 3.39
MM1 pKa = 7.66LFRR4 pKa = 11.84RR5 pKa = 11.84THH7 pKa = 5.36LLFRR11 pKa = 11.84RR12 pKa = 11.84THH14 pKa = 5.49LLFRR18 pKa = 11.84RR19 pKa = 11.84THH21 pKa = 5.49LLFRR25 pKa = 11.84RR26 pKa = 11.84THH28 pKa = 5.49LLFRR32 pKa = 11.84RR33 pKa = 11.84THH35 pKa = 5.49LLFRR39 pKa = 11.84RR40 pKa = 11.84THH42 pKa = 5.49LLFRR46 pKa = 11.84RR47 pKa = 11.84THH49 pKa = 5.49LLFRR53 pKa = 11.84RR54 pKa = 11.84THH56 pKa = 5.49LLFRR60 pKa = 11.84RR61 pKa = 11.84THH63 pKa = 5.49LLFRR67 pKa = 11.84RR68 pKa = 11.84THH70 pKa = 5.36LLFRR74 pKa = 11.84RR75 pKa = 11.84VHH77 pKa = 5.46NAPYY81 pKa = 10.16KK82 pKa = 7.48ITRR85 pKa = 11.84RR86 pKa = 11.84VEE88 pKa = 3.95TYY90 pKa = 8.97TGHH93 pKa = 6.0NTVQLSPKK101 pKa = 9.33TLVKK105 pKa = 9.75TRR107 pKa = 11.84NFASLQVV114 pKa = 3.39
Molecular weight: 14.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2503695 |
37 |
4984 |
244.7 |
27.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.813 ± 0.03 | 1.021 ± 0.01 |
4.882 ± 0.025 | 6.268 ± 0.03 |
3.997 ± 0.02 | 6.538 ± 0.038 |
1.773 ± 0.015 | 6.911 ± 0.024 |
5.175 ± 0.026 | 10.891 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.795 ± 0.013 | 4.682 ± 0.031 |
4.54 ± 0.024 | 5.372 ± 0.025 |
5.054 ± 0.029 | 6.691 ± 0.026 |
5.704 ± 0.024 | 6.438 ± 0.022 |
1.405 ± 0.011 | 3.047 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
