
Drosophila busckii rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
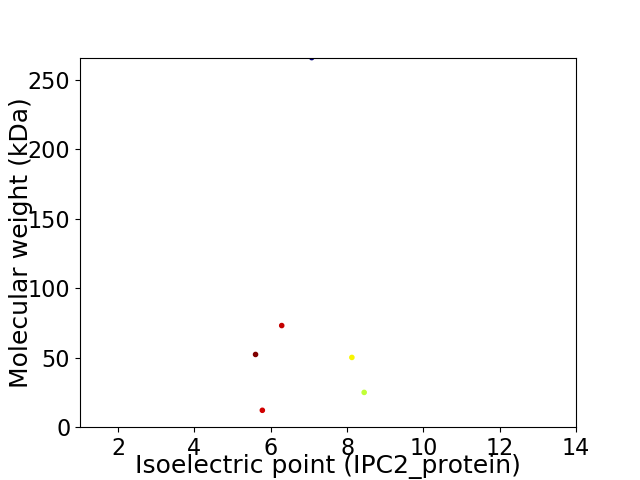
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A140D8L2|A0A140D8L2_9RHAB Polymerase-associated protein OS=Drosophila busckii rhabdovirus OX=1802935 PE=4 SV=1
MM1 pKa = 7.38AHH3 pKa = 6.91SSSQQISPRR12 pKa = 11.84DD13 pKa = 3.73VQILTDD19 pKa = 3.46GHH21 pKa = 5.89SVVAQMKK28 pKa = 9.22GISSVGEE35 pKa = 4.07SEE37 pKa = 4.73IQDD40 pKa = 4.15LPWTEE45 pKa = 4.16STFGEE50 pKa = 4.42LLDD53 pKa = 3.58MYY55 pKa = 9.82GWKK58 pKa = 9.4VHH60 pKa = 6.92PEE62 pKa = 3.68DD63 pKa = 3.91TVINGAASSILNWYY77 pKa = 8.56MGGPTKK83 pKa = 10.83NLLQSIEE90 pKa = 4.08VVIAGMTVSGSGTPLITIKK109 pKa = 10.95DD110 pKa = 3.55RR111 pKa = 11.84GTKK114 pKa = 8.26STHH117 pKa = 4.96KK118 pKa = 9.22WPSIVIPKK126 pKa = 10.28RR127 pKa = 11.84LDD129 pKa = 3.41GDD131 pKa = 4.03QLAKK135 pKa = 9.94WKK137 pKa = 9.34PALEE141 pKa = 4.29NLDD144 pKa = 3.7QEE146 pKa = 4.75ISIAPPALPTSVEE159 pKa = 4.13GAEE162 pKa = 4.34TTRR165 pKa = 11.84MATSQTEE172 pKa = 3.63AGKK175 pKa = 9.6WYY177 pKa = 10.43DD178 pKa = 4.33FMWDD182 pKa = 3.73GLNTAITAEE191 pKa = 4.28DD192 pKa = 3.78PKK194 pKa = 11.16ADD196 pKa = 4.39EE197 pKa = 4.3IRR199 pKa = 11.84KK200 pKa = 9.14KK201 pKa = 10.38RR202 pKa = 11.84QFIINYY208 pKa = 8.66VGYY211 pKa = 10.18IGLNLLRR218 pKa = 11.84GVAKK222 pKa = 10.76GSTTVIRR229 pKa = 11.84ALEE232 pKa = 4.14KK233 pKa = 10.88NLVEE237 pKa = 5.25RR238 pKa = 11.84IPHH241 pKa = 5.66FWPTEE246 pKa = 3.96FQVPPPPMGAQSAIQEE262 pKa = 4.22IEE264 pKa = 3.88NMLEE268 pKa = 4.16RR269 pKa = 11.84GSEE272 pKa = 4.04GCRR275 pKa = 11.84RR276 pKa = 11.84LIVNMIPDD284 pKa = 4.02TLSTEE289 pKa = 4.08PAKK292 pKa = 10.73KK293 pKa = 10.07GILMSGCMKK302 pKa = 10.49SLSDD306 pKa = 3.56SGLGALHH313 pKa = 6.65WVYY316 pKa = 10.62EE317 pKa = 4.03AAAYY321 pKa = 7.66LTVEE325 pKa = 4.87PGDD328 pKa = 3.83LMEE331 pKa = 4.24TMYY334 pKa = 10.63YY335 pKa = 10.82GPFIASSQEE344 pKa = 3.77VAGFLSSMHH353 pKa = 6.44GLPKK357 pKa = 8.05EE358 pKa = 4.37WKK360 pKa = 7.15WARR363 pKa = 11.84VINEE367 pKa = 4.15GYY369 pKa = 9.83ISKK372 pKa = 10.78LSVRR376 pKa = 11.84NNKK379 pKa = 9.9KK380 pKa = 10.17YY381 pKa = 10.68VATLVALSCGMSPDD395 pKa = 3.56SPIWAIPSLAINNLIKK411 pKa = 10.6RR412 pKa = 11.84EE413 pKa = 4.02ALDD416 pKa = 3.61NAQAIYY422 pKa = 10.96SEE424 pKa = 4.37LTDD427 pKa = 3.44TRR429 pKa = 11.84NYY431 pKa = 10.62AAGSAKK437 pKa = 10.11SAAVVAALSRR447 pKa = 11.84IRR449 pKa = 11.84TRR451 pKa = 11.84DD452 pKa = 3.53PLEE455 pKa = 4.77GIPTAVPCAPVSKK468 pKa = 10.47CPPTSDD474 pKa = 3.21GFGRR478 pKa = 11.84EE479 pKa = 4.1YY480 pKa = 11.41
MM1 pKa = 7.38AHH3 pKa = 6.91SSSQQISPRR12 pKa = 11.84DD13 pKa = 3.73VQILTDD19 pKa = 3.46GHH21 pKa = 5.89SVVAQMKK28 pKa = 9.22GISSVGEE35 pKa = 4.07SEE37 pKa = 4.73IQDD40 pKa = 4.15LPWTEE45 pKa = 4.16STFGEE50 pKa = 4.42LLDD53 pKa = 3.58MYY55 pKa = 9.82GWKK58 pKa = 9.4VHH60 pKa = 6.92PEE62 pKa = 3.68DD63 pKa = 3.91TVINGAASSILNWYY77 pKa = 8.56MGGPTKK83 pKa = 10.83NLLQSIEE90 pKa = 4.08VVIAGMTVSGSGTPLITIKK109 pKa = 10.95DD110 pKa = 3.55RR111 pKa = 11.84GTKK114 pKa = 8.26STHH117 pKa = 4.96KK118 pKa = 9.22WPSIVIPKK126 pKa = 10.28RR127 pKa = 11.84LDD129 pKa = 3.41GDD131 pKa = 4.03QLAKK135 pKa = 9.94WKK137 pKa = 9.34PALEE141 pKa = 4.29NLDD144 pKa = 3.7QEE146 pKa = 4.75ISIAPPALPTSVEE159 pKa = 4.13GAEE162 pKa = 4.34TTRR165 pKa = 11.84MATSQTEE172 pKa = 3.63AGKK175 pKa = 9.6WYY177 pKa = 10.43DD178 pKa = 4.33FMWDD182 pKa = 3.73GLNTAITAEE191 pKa = 4.28DD192 pKa = 3.78PKK194 pKa = 11.16ADD196 pKa = 4.39EE197 pKa = 4.3IRR199 pKa = 11.84KK200 pKa = 9.14KK201 pKa = 10.38RR202 pKa = 11.84QFIINYY208 pKa = 8.66VGYY211 pKa = 10.18IGLNLLRR218 pKa = 11.84GVAKK222 pKa = 10.76GSTTVIRR229 pKa = 11.84ALEE232 pKa = 4.14KK233 pKa = 10.88NLVEE237 pKa = 5.25RR238 pKa = 11.84IPHH241 pKa = 5.66FWPTEE246 pKa = 3.96FQVPPPPMGAQSAIQEE262 pKa = 4.22IEE264 pKa = 3.88NMLEE268 pKa = 4.16RR269 pKa = 11.84GSEE272 pKa = 4.04GCRR275 pKa = 11.84RR276 pKa = 11.84LIVNMIPDD284 pKa = 4.02TLSTEE289 pKa = 4.08PAKK292 pKa = 10.73KK293 pKa = 10.07GILMSGCMKK302 pKa = 10.49SLSDD306 pKa = 3.56SGLGALHH313 pKa = 6.65WVYY316 pKa = 10.62EE317 pKa = 4.03AAAYY321 pKa = 7.66LTVEE325 pKa = 4.87PGDD328 pKa = 3.83LMEE331 pKa = 4.24TMYY334 pKa = 10.63YY335 pKa = 10.82GPFIASSQEE344 pKa = 3.77VAGFLSSMHH353 pKa = 6.44GLPKK357 pKa = 8.05EE358 pKa = 4.37WKK360 pKa = 7.15WARR363 pKa = 11.84VINEE367 pKa = 4.15GYY369 pKa = 9.83ISKK372 pKa = 10.78LSVRR376 pKa = 11.84NNKK379 pKa = 9.9KK380 pKa = 10.17YY381 pKa = 10.68VATLVALSCGMSPDD395 pKa = 3.56SPIWAIPSLAINNLIKK411 pKa = 10.6RR412 pKa = 11.84EE413 pKa = 4.02ALDD416 pKa = 3.61NAQAIYY422 pKa = 10.96SEE424 pKa = 4.37LTDD427 pKa = 3.44TRR429 pKa = 11.84NYY431 pKa = 10.62AAGSAKK437 pKa = 10.11SAAVVAALSRR447 pKa = 11.84IRR449 pKa = 11.84TRR451 pKa = 11.84DD452 pKa = 3.53PLEE455 pKa = 4.77GIPTAVPCAPVSKK468 pKa = 10.47CPPTSDD474 pKa = 3.21GFGRR478 pKa = 11.84EE479 pKa = 4.1YY480 pKa = 11.41
Molecular weight: 52.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140D8L4|A0A140D8L4_9RHAB Glycoprotein OS=Drosophila busckii rhabdovirus OX=1802935 GN=G PE=4 SV=1
MM1 pKa = 7.56AKK3 pKa = 9.92INHH6 pKa = 7.11DD7 pKa = 3.47YY8 pKa = 10.92HH9 pKa = 6.72NRR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 6.07FKK15 pKa = 8.37TTVQVDD21 pKa = 3.19ILLWGTSEE29 pKa = 5.16IDD31 pKa = 3.36PEE33 pKa = 4.46LLKK36 pKa = 11.06KK37 pKa = 9.82MCACIIWEE45 pKa = 4.7GYY47 pKa = 9.43KK48 pKa = 10.84GIVEE52 pKa = 4.81DD53 pKa = 4.37PMMWKK58 pKa = 9.35QASQALYY65 pKa = 10.8SKK67 pKa = 10.45IRR69 pKa = 11.84DD70 pKa = 3.43KK71 pKa = 11.49SRR73 pKa = 11.84WSPVLSDD80 pKa = 4.36LNHH83 pKa = 6.27HH84 pKa = 6.65AVLEE88 pKa = 4.28ATWFSLTPRR97 pKa = 11.84IIAFPDD103 pKa = 3.35DD104 pKa = 3.94HH105 pKa = 6.79KK106 pKa = 11.47VSYY109 pKa = 10.89SRR111 pKa = 11.84LFNFRR116 pKa = 11.84APGGKK121 pKa = 8.87ILSVDD126 pKa = 3.67YY127 pKa = 9.93KK128 pKa = 10.49IHH130 pKa = 6.5MIHH133 pKa = 6.67NIWDD137 pKa = 3.25KK138 pKa = 11.29RR139 pKa = 11.84EE140 pKa = 3.84YY141 pKa = 10.97GNLVMRR147 pKa = 11.84GYY149 pKa = 9.0TCLEE153 pKa = 3.85QLGLTPRR160 pKa = 11.84IGGVISLAIEE170 pKa = 4.02KK171 pKa = 10.0MIGTVVLDD179 pKa = 4.0PYY181 pKa = 11.64KK182 pKa = 10.05MVSLMEE188 pKa = 4.08MAPLIEE194 pKa = 4.12QTEE197 pKa = 4.65VIDD200 pKa = 4.1TPSGPKK206 pKa = 9.56LKK208 pKa = 10.52KK209 pKa = 10.46GFLSQFRR216 pKa = 11.84KK217 pKa = 10.2
MM1 pKa = 7.56AKK3 pKa = 9.92INHH6 pKa = 7.11DD7 pKa = 3.47YY8 pKa = 10.92HH9 pKa = 6.72NRR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 6.07FKK15 pKa = 8.37TTVQVDD21 pKa = 3.19ILLWGTSEE29 pKa = 5.16IDD31 pKa = 3.36PEE33 pKa = 4.46LLKK36 pKa = 11.06KK37 pKa = 9.82MCACIIWEE45 pKa = 4.7GYY47 pKa = 9.43KK48 pKa = 10.84GIVEE52 pKa = 4.81DD53 pKa = 4.37PMMWKK58 pKa = 9.35QASQALYY65 pKa = 10.8SKK67 pKa = 10.45IRR69 pKa = 11.84DD70 pKa = 3.43KK71 pKa = 11.49SRR73 pKa = 11.84WSPVLSDD80 pKa = 4.36LNHH83 pKa = 6.27HH84 pKa = 6.65AVLEE88 pKa = 4.28ATWFSLTPRR97 pKa = 11.84IIAFPDD103 pKa = 3.35DD104 pKa = 3.94HH105 pKa = 6.79KK106 pKa = 11.47VSYY109 pKa = 10.89SRR111 pKa = 11.84LFNFRR116 pKa = 11.84APGGKK121 pKa = 8.87ILSVDD126 pKa = 3.67YY127 pKa = 9.93KK128 pKa = 10.49IHH130 pKa = 6.5MIHH133 pKa = 6.67NIWDD137 pKa = 3.25KK138 pKa = 11.29RR139 pKa = 11.84EE140 pKa = 3.84YY141 pKa = 10.97GNLVMRR147 pKa = 11.84GYY149 pKa = 9.0TCLEE153 pKa = 3.85QLGLTPRR160 pKa = 11.84IGGVISLAIEE170 pKa = 4.02KK171 pKa = 10.0MIGTVVLDD179 pKa = 4.0PYY181 pKa = 11.64KK182 pKa = 10.05MVSLMEE188 pKa = 4.08MAPLIEE194 pKa = 4.12QTEE197 pKa = 4.65VIDD200 pKa = 4.1TPSGPKK206 pKa = 9.56LKK208 pKa = 10.52KK209 pKa = 10.46GFLSQFRR216 pKa = 11.84KK217 pKa = 10.2
Molecular weight: 25.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4214 |
111 |
2327 |
702.3 |
79.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.865 ± 0.696 | 1.732 ± 0.21 |
4.722 ± 0.172 | 5.956 ± 0.379 |
4.058 ± 0.669 | 5.434 ± 0.52 |
2.492 ± 0.222 | 7.143 ± 0.194 |
5.766 ± 0.465 | 9.991 ± 0.627 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.539 ± 0.244 | 4.746 ± 0.337 |
6.407 ± 0.401 | 3.821 ± 0.401 |
5.173 ± 0.19 | 8.353 ± 0.416 |
6.122 ± 0.274 | 5.15 ± 0.308 |
1.875 ± 0.191 | 3.654 ± 0.246 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
