
Actinomyces sp. 2119
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Actinomyces; unclassified Actinomyces
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
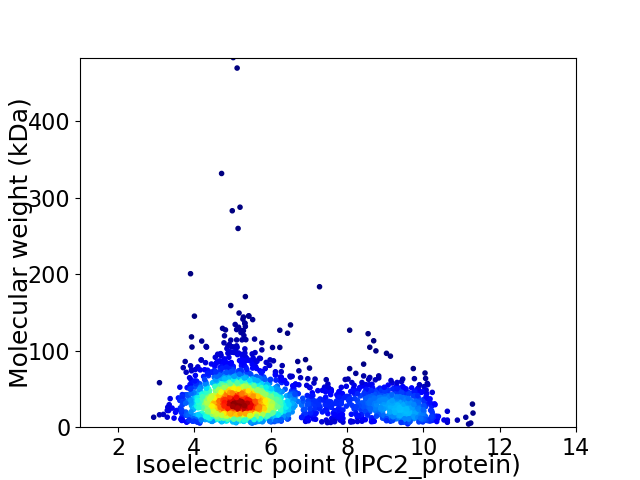
Virtual 2D-PAGE plot for 2334 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A418UR61|A0A418UR61_9ACTO Copper-binding protein OS=Actinomyces sp. 2119 OX=2321393 GN=D4740_10715 PE=4 SV=1
MM1 pKa = 7.29LRR3 pKa = 11.84IGNRR7 pKa = 11.84VTDD10 pKa = 3.75TFTVGNRR17 pKa = 11.84TSTALGLTNTADD29 pKa = 3.84YY30 pKa = 11.06KK31 pKa = 10.87PGTLAVTKK39 pKa = 9.79TVTGLEE45 pKa = 3.92AAAPRR50 pKa = 11.84TYY52 pKa = 10.71SYY54 pKa = 10.53TYY56 pKa = 9.36TCGEE60 pKa = 4.06VTGTISNVPGDD71 pKa = 4.18GTPVEE76 pKa = 4.61AGATIPAGTEE86 pKa = 3.87CTVTEE91 pKa = 4.57DD92 pKa = 2.93VDD94 pKa = 3.6AAAMDD99 pKa = 4.97GYY101 pKa = 11.19DD102 pKa = 4.27LAAPDD107 pKa = 4.44PVTVTVVPSEE117 pKa = 4.15EE118 pKa = 4.25AVTEE122 pKa = 4.02AAFTNSYY129 pKa = 8.72TRR131 pKa = 11.84HH132 pKa = 5.79TGTFSVAKK140 pKa = 9.7QVGPEE145 pKa = 4.13DD146 pKa = 3.82VPFLADD152 pKa = 3.68TFDD155 pKa = 3.23VDD157 pKa = 4.06YY158 pKa = 10.44TCTQPEE164 pKa = 4.37GTEE167 pKa = 3.99VSGTLQVTGGAPAVNGPTLPVGTTCSVSEE196 pKa = 4.39PEE198 pKa = 4.18EE199 pKa = 3.91TTQRR203 pKa = 11.84DD204 pKa = 4.2GYY206 pKa = 10.93NVSTLIRR213 pKa = 11.84VDD215 pKa = 3.26GTEE218 pKa = 3.96GSRR221 pKa = 11.84LTITRR226 pKa = 11.84DD227 pKa = 3.16TTSEE231 pKa = 4.05VSVVNAYY238 pKa = 7.48TALTGGFQISKK249 pKa = 10.1EE250 pKa = 4.14VTGDD254 pKa = 3.38GAGLADD260 pKa = 3.48SQTYY264 pKa = 9.16VFTYY268 pKa = 9.48TCTGVNGTTTTDD280 pKa = 3.16TVEE283 pKa = 4.21LVSGATTAVTDD294 pKa = 4.02VPVGSCTVNEE304 pKa = 4.29ADD306 pKa = 4.75ASVDD310 pKa = 3.6GADD313 pKa = 4.65LTTTWSVDD321 pKa = 3.09GEE323 pKa = 4.37EE324 pKa = 4.37TDD326 pKa = 4.36GEE328 pKa = 4.8VTFDD332 pKa = 3.55VADD335 pKa = 4.01GATAVVQATNDD346 pKa = 3.66YY347 pKa = 8.36TVHH350 pKa = 6.92RR351 pKa = 11.84GGFSVAKK358 pKa = 9.11EE359 pKa = 3.86VTGAEE364 pKa = 4.14EE365 pKa = 4.43ADD367 pKa = 3.76LSAKK371 pKa = 10.11EE372 pKa = 4.04FGVTYY377 pKa = 9.53TCTDD381 pKa = 2.72GSTGVLAVGADD392 pKa = 3.94GQSVSGPQVPVGTEE406 pKa = 3.94CTVSEE411 pKa = 4.5DD412 pKa = 3.48LASASLAGYY421 pKa = 7.48TLTAPEE427 pKa = 4.32DD428 pKa = 3.44QAVTVEE434 pKa = 4.07ARR436 pKa = 11.84GQEE439 pKa = 4.15TALTLSNAYY448 pKa = 8.27TRR450 pKa = 11.84QTGSFAVSKK459 pKa = 9.71TVDD462 pKa = 2.97GDD464 pKa = 3.72GAARR468 pKa = 11.84APQSFSFAYY477 pKa = 8.98TCTGVDD483 pKa = 3.37GTTTTGTVPAADD495 pKa = 4.01ADD497 pKa = 4.29SAVVTDD503 pKa = 4.09VPVGRR508 pKa = 11.84CTISEE513 pKa = 3.9EE514 pKa = 4.14DD515 pKa = 3.61ASVEE519 pKa = 4.11GTDD522 pKa = 5.31LLTQTVVDD530 pKa = 4.43GDD532 pKa = 3.95PVITEE537 pKa = 4.12EE538 pKa = 4.95ASLTVTDD545 pKa = 3.97GGMVAVVVTNTYY557 pKa = 8.24TLHH560 pKa = 6.7RR561 pKa = 11.84GSFSVTKK568 pKa = 10.36VVEE571 pKa = 3.99GLEE574 pKa = 4.55DD575 pKa = 4.07DD576 pKa = 5.2SEE578 pKa = 4.66DD579 pKa = 4.79AGDD582 pKa = 4.78PDD584 pKa = 3.86AQAAPRR590 pKa = 11.84DD591 pKa = 3.71YY592 pKa = 11.08LFTYY596 pKa = 9.39TCTLPEE602 pKa = 4.47DD603 pKa = 3.78AADD606 pKa = 4.06AEE608 pKa = 4.71GGADD612 pKa = 3.3GGVVSDD618 pKa = 4.32QVTVPAGMTVTSPALPVGTEE638 pKa = 4.11CTVSEE643 pKa = 4.43DD644 pKa = 3.53AEE646 pKa = 4.09AAAVDD651 pKa = 5.34GYY653 pKa = 11.52ALEE656 pKa = 4.4PAEE659 pKa = 4.46DD660 pKa = 3.55QAVTVSTQDD669 pKa = 3.1TVEE672 pKa = 4.42EE673 pKa = 4.17MTFTNTYY680 pKa = 6.96TRR682 pKa = 11.84EE683 pKa = 4.1VEE685 pKa = 4.2PTPLPSEE692 pKa = 4.39SPAPQPSGEE701 pKa = 4.22PSTGPSAVVTSGPATPSSEE720 pKa = 4.39VPASASPSPGGSSVTGSLARR740 pKa = 11.84TGAAILLPAVAALAAIGGGSLLLRR764 pKa = 11.84RR765 pKa = 11.84RR766 pKa = 11.84RR767 pKa = 11.84QQ768 pKa = 3.05
MM1 pKa = 7.29LRR3 pKa = 11.84IGNRR7 pKa = 11.84VTDD10 pKa = 3.75TFTVGNRR17 pKa = 11.84TSTALGLTNTADD29 pKa = 3.84YY30 pKa = 11.06KK31 pKa = 10.87PGTLAVTKK39 pKa = 9.79TVTGLEE45 pKa = 3.92AAAPRR50 pKa = 11.84TYY52 pKa = 10.71SYY54 pKa = 10.53TYY56 pKa = 9.36TCGEE60 pKa = 4.06VTGTISNVPGDD71 pKa = 4.18GTPVEE76 pKa = 4.61AGATIPAGTEE86 pKa = 3.87CTVTEE91 pKa = 4.57DD92 pKa = 2.93VDD94 pKa = 3.6AAAMDD99 pKa = 4.97GYY101 pKa = 11.19DD102 pKa = 4.27LAAPDD107 pKa = 4.44PVTVTVVPSEE117 pKa = 4.15EE118 pKa = 4.25AVTEE122 pKa = 4.02AAFTNSYY129 pKa = 8.72TRR131 pKa = 11.84HH132 pKa = 5.79TGTFSVAKK140 pKa = 9.7QVGPEE145 pKa = 4.13DD146 pKa = 3.82VPFLADD152 pKa = 3.68TFDD155 pKa = 3.23VDD157 pKa = 4.06YY158 pKa = 10.44TCTQPEE164 pKa = 4.37GTEE167 pKa = 3.99VSGTLQVTGGAPAVNGPTLPVGTTCSVSEE196 pKa = 4.39PEE198 pKa = 4.18EE199 pKa = 3.91TTQRR203 pKa = 11.84DD204 pKa = 4.2GYY206 pKa = 10.93NVSTLIRR213 pKa = 11.84VDD215 pKa = 3.26GTEE218 pKa = 3.96GSRR221 pKa = 11.84LTITRR226 pKa = 11.84DD227 pKa = 3.16TTSEE231 pKa = 4.05VSVVNAYY238 pKa = 7.48TALTGGFQISKK249 pKa = 10.1EE250 pKa = 4.14VTGDD254 pKa = 3.38GAGLADD260 pKa = 3.48SQTYY264 pKa = 9.16VFTYY268 pKa = 9.48TCTGVNGTTTTDD280 pKa = 3.16TVEE283 pKa = 4.21LVSGATTAVTDD294 pKa = 4.02VPVGSCTVNEE304 pKa = 4.29ADD306 pKa = 4.75ASVDD310 pKa = 3.6GADD313 pKa = 4.65LTTTWSVDD321 pKa = 3.09GEE323 pKa = 4.37EE324 pKa = 4.37TDD326 pKa = 4.36GEE328 pKa = 4.8VTFDD332 pKa = 3.55VADD335 pKa = 4.01GATAVVQATNDD346 pKa = 3.66YY347 pKa = 8.36TVHH350 pKa = 6.92RR351 pKa = 11.84GGFSVAKK358 pKa = 9.11EE359 pKa = 3.86VTGAEE364 pKa = 4.14EE365 pKa = 4.43ADD367 pKa = 3.76LSAKK371 pKa = 10.11EE372 pKa = 4.04FGVTYY377 pKa = 9.53TCTDD381 pKa = 2.72GSTGVLAVGADD392 pKa = 3.94GQSVSGPQVPVGTEE406 pKa = 3.94CTVSEE411 pKa = 4.5DD412 pKa = 3.48LASASLAGYY421 pKa = 7.48TLTAPEE427 pKa = 4.32DD428 pKa = 3.44QAVTVEE434 pKa = 4.07ARR436 pKa = 11.84GQEE439 pKa = 4.15TALTLSNAYY448 pKa = 8.27TRR450 pKa = 11.84QTGSFAVSKK459 pKa = 9.71TVDD462 pKa = 2.97GDD464 pKa = 3.72GAARR468 pKa = 11.84APQSFSFAYY477 pKa = 8.98TCTGVDD483 pKa = 3.37GTTTTGTVPAADD495 pKa = 4.01ADD497 pKa = 4.29SAVVTDD503 pKa = 4.09VPVGRR508 pKa = 11.84CTISEE513 pKa = 3.9EE514 pKa = 4.14DD515 pKa = 3.61ASVEE519 pKa = 4.11GTDD522 pKa = 5.31LLTQTVVDD530 pKa = 4.43GDD532 pKa = 3.95PVITEE537 pKa = 4.12EE538 pKa = 4.95ASLTVTDD545 pKa = 3.97GGMVAVVVTNTYY557 pKa = 8.24TLHH560 pKa = 6.7RR561 pKa = 11.84GSFSVTKK568 pKa = 10.36VVEE571 pKa = 3.99GLEE574 pKa = 4.55DD575 pKa = 4.07DD576 pKa = 5.2SEE578 pKa = 4.66DD579 pKa = 4.79AGDD582 pKa = 4.78PDD584 pKa = 3.86AQAAPRR590 pKa = 11.84DD591 pKa = 3.71YY592 pKa = 11.08LFTYY596 pKa = 9.39TCTLPEE602 pKa = 4.47DD603 pKa = 3.78AADD606 pKa = 4.06AEE608 pKa = 4.71GGADD612 pKa = 3.3GGVVSDD618 pKa = 4.32QVTVPAGMTVTSPALPVGTEE638 pKa = 4.11CTVSEE643 pKa = 4.43DD644 pKa = 3.53AEE646 pKa = 4.09AAAVDD651 pKa = 5.34GYY653 pKa = 11.52ALEE656 pKa = 4.4PAEE659 pKa = 4.46DD660 pKa = 3.55QAVTVSTQDD669 pKa = 3.1TVEE672 pKa = 4.42EE673 pKa = 4.17MTFTNTYY680 pKa = 6.96TRR682 pKa = 11.84EE683 pKa = 4.1VEE685 pKa = 4.2PTPLPSEE692 pKa = 4.39SPAPQPSGEE701 pKa = 4.22PSTGPSAVVTSGPATPSSEE720 pKa = 4.39VPASASPSPGGSSVTGSLARR740 pKa = 11.84TGAAILLPAVAALAAIGGGSLLLRR764 pKa = 11.84RR765 pKa = 11.84RR766 pKa = 11.84RR767 pKa = 11.84QQ768 pKa = 3.05
Molecular weight: 77.75 kDa
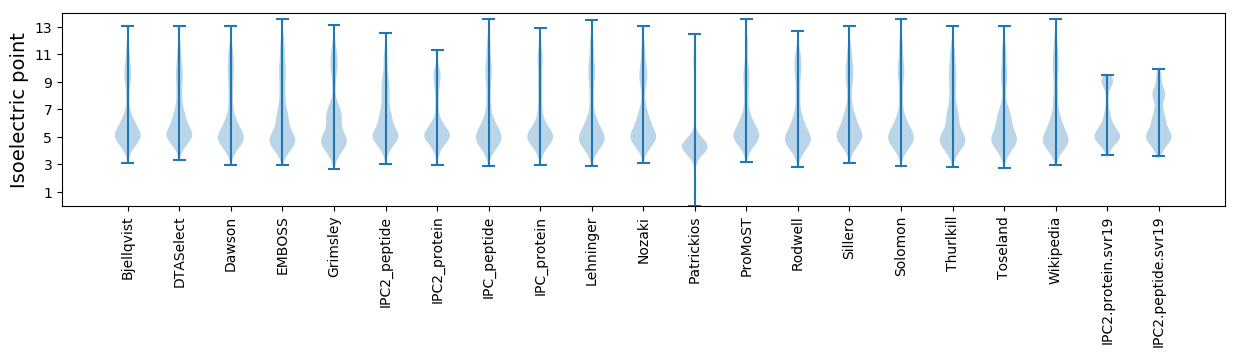
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A418UQ06|A0A418UQ06_9ACTO TatD family deoxyribonuclease OS=Actinomyces sp. 2119 OX=2321393 GN=D4740_12250 PE=4 SV=1
MM1 pKa = 7.32LLVEE5 pKa = 4.58RR6 pKa = 11.84VLVSLLPTLLKK17 pKa = 10.53PLGHH21 pKa = 6.63RR22 pKa = 11.84PITRR26 pKa = 11.84GATITVTVRR35 pKa = 11.84ATAATLGRR43 pKa = 11.84AVPATTTAGATRR55 pKa = 11.84AVTTTTGTVSTVSRR69 pKa = 11.84VAGRR73 pKa = 11.84GLTVSPVSPVAARR86 pKa = 11.84RR87 pKa = 11.84GIRR90 pKa = 11.84ISTLGVGAVTGRR102 pKa = 11.84GLTVSTLSAVTASRR116 pKa = 11.84GLTIGALTTAVTRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84LRR133 pKa = 11.84TITISPVAPRR143 pKa = 11.84RR144 pKa = 11.84GTRR147 pKa = 11.84LSRR150 pKa = 11.84VTISTVAGRR159 pKa = 11.84GARR162 pKa = 11.84LSGTVSVSLVATPRR176 pKa = 11.84TRR178 pKa = 11.84LSRR181 pKa = 3.68
MM1 pKa = 7.32LLVEE5 pKa = 4.58RR6 pKa = 11.84VLVSLLPTLLKK17 pKa = 10.53PLGHH21 pKa = 6.63RR22 pKa = 11.84PITRR26 pKa = 11.84GATITVTVRR35 pKa = 11.84ATAATLGRR43 pKa = 11.84AVPATTTAGATRR55 pKa = 11.84AVTTTTGTVSTVSRR69 pKa = 11.84VAGRR73 pKa = 11.84GLTVSPVSPVAARR86 pKa = 11.84RR87 pKa = 11.84GIRR90 pKa = 11.84ISTLGVGAVTGRR102 pKa = 11.84GLTVSTLSAVTASRR116 pKa = 11.84GLTIGALTTAVTRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84LRR133 pKa = 11.84TITISPVAPRR143 pKa = 11.84RR144 pKa = 11.84GTRR147 pKa = 11.84LSRR150 pKa = 11.84VTISTVAGRR159 pKa = 11.84GARR162 pKa = 11.84LSGTVSVSLVATPRR176 pKa = 11.84TRR178 pKa = 11.84LSRR181 pKa = 3.68
Molecular weight: 18.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
835853 |
32 |
4575 |
358.1 |
38.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.126 ± 0.06 | 0.809 ± 0.016 |
5.764 ± 0.039 | 5.709 ± 0.04 |
2.316 ± 0.029 | 9.352 ± 0.039 |
2.052 ± 0.022 | 2.88 ± 0.035 |
1.327 ± 0.021 | 10.323 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.855 ± 0.022 | 1.548 ± 0.022 |
5.767 ± 0.05 | 3.448 ± 0.023 |
7.807 ± 0.053 | 6.373 ± 0.038 |
6.378 ± 0.05 | 9.727 ± 0.048 |
1.463 ± 0.02 | 1.977 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
