
Avian metaavulavirus 8
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Metaavulavirus
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
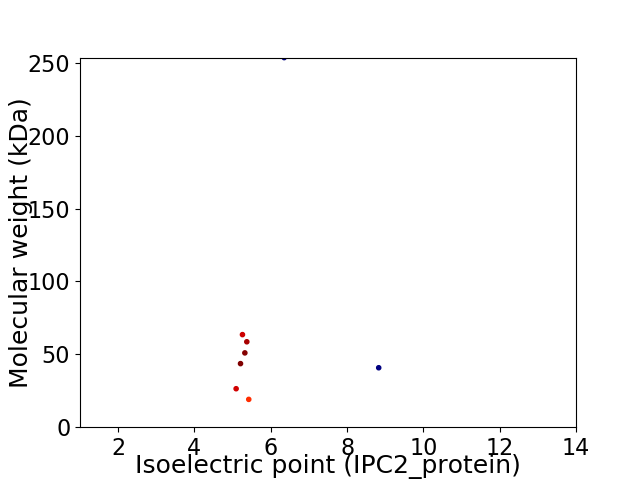
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|C5I0V2|C5I0V2_9MONO Phosphoprotein OS=Avian metaavulavirus 8 OX=2560318 GN=P PE=4 SV=1
MM1 pKa = 7.39SNIASSLEE9 pKa = 4.09NIVEE13 pKa = 3.88QDD15 pKa = 3.05SRR17 pKa = 11.84KK18 pKa = 6.29TTWRR22 pKa = 11.84AIFRR26 pKa = 11.84WSVLLITTGCLALSIVSIVQIGNLKK51 pKa = 9.78IPSVGDD57 pKa = 3.38LADD60 pKa = 3.74EE61 pKa = 4.38VVTPLKK67 pKa = 8.49TTLSDD72 pKa = 3.5TLRR75 pKa = 11.84NPINQINDD83 pKa = 2.72IFRR86 pKa = 11.84IVALDD91 pKa = 3.39IPLQVTSIQKK101 pKa = 10.27DD102 pKa = 3.48LASQFSMLIDD112 pKa = 3.39SLNAIKK118 pKa = 10.61LGNGTNLIIPTSDD131 pKa = 2.77KK132 pKa = 10.71EE133 pKa = 4.18YY134 pKa = 10.99AGGIGNPVFTVDD146 pKa = 3.25AGGSIGFKK154 pKa = 9.57QFSLIEE160 pKa = 4.29HH161 pKa = 7.17PSFIAGPTTTRR172 pKa = 11.84GCTRR176 pKa = 11.84IPTFHH181 pKa = 6.76MSEE184 pKa = 4.21SHH186 pKa = 5.05WCYY189 pKa = 9.94SHH191 pKa = 7.04NIIAAGCQDD200 pKa = 3.49ASASSMYY207 pKa = 10.46ISMGVLHH214 pKa = 6.54VSSSGTPIFLTTASEE229 pKa = 4.59LIDD232 pKa = 4.71DD233 pKa = 4.47GVNRR237 pKa = 11.84KK238 pKa = 9.16SCSIVATQFGCDD250 pKa = 3.31ILCSIVIEE258 pKa = 4.95KK259 pKa = 10.73EE260 pKa = 3.82GDD262 pKa = 3.87DD263 pKa = 3.97YY264 pKa = 11.52WSDD267 pKa = 3.24TPTPMRR273 pKa = 11.84HH274 pKa = 5.49GRR276 pKa = 11.84FSFNGSFVEE285 pKa = 4.39TEE287 pKa = 4.37LPVSSMFSSFSANYY301 pKa = 8.0PAVGSGEE308 pKa = 4.07IVKK311 pKa = 10.47DD312 pKa = 3.88RR313 pKa = 11.84ILFPIYY319 pKa = 10.55GGIKK323 pKa = 7.85QTSPEE328 pKa = 3.75FTEE331 pKa = 4.16LVKK334 pKa = 11.22YY335 pKa = 10.5GLFVSTPTTVCQSSWTYY352 pKa = 11.11DD353 pKa = 3.39QVKK356 pKa = 9.54AAYY359 pKa = 9.45RR360 pKa = 11.84PDD362 pKa = 4.15YY363 pKa = 10.62ISGRR367 pKa = 11.84FWAQVILSCALDD379 pKa = 3.78AVDD382 pKa = 5.94LSSCIVKK389 pKa = 10.13IMNSSTVMMAAEE401 pKa = 4.06GRR403 pKa = 11.84IIKK406 pKa = 10.18IGIDD410 pKa = 3.03YY411 pKa = 9.78FYY413 pKa = 10.53YY414 pKa = 10.3QRR416 pKa = 11.84SSSWWPLAFVTKK428 pKa = 10.57LDD430 pKa = 3.92PQEE433 pKa = 4.79LADD436 pKa = 4.56TNSIWLTNSIPIPQSKK452 pKa = 9.59FPRR455 pKa = 11.84PSYY458 pKa = 10.67SEE460 pKa = 4.17NYY462 pKa = 6.58CTKK465 pKa = 10.29PAVCPATCVTGVYY478 pKa = 10.43SDD480 pKa = 3.44IWPLTSSSSLPSIIWIGQYY499 pKa = 10.79LDD501 pKa = 3.82APVGRR506 pKa = 11.84TYY508 pKa = 11.06PRR510 pKa = 11.84FGIANQSHH518 pKa = 6.99WYY520 pKa = 8.34LQEE523 pKa = 5.89DD524 pKa = 3.92ILPTSTASAYY534 pKa = 8.49STTTCFKK541 pKa = 8.14NTARR545 pKa = 11.84NRR547 pKa = 11.84VFCVTIAEE555 pKa = 4.31FADD558 pKa = 3.97GLFGEE563 pKa = 4.55YY564 pKa = 9.99RR565 pKa = 11.84ITPQLYY571 pKa = 9.5EE572 pKa = 3.99LVRR575 pKa = 11.84NNN577 pKa = 3.26
MM1 pKa = 7.39SNIASSLEE9 pKa = 4.09NIVEE13 pKa = 3.88QDD15 pKa = 3.05SRR17 pKa = 11.84KK18 pKa = 6.29TTWRR22 pKa = 11.84AIFRR26 pKa = 11.84WSVLLITTGCLALSIVSIVQIGNLKK51 pKa = 9.78IPSVGDD57 pKa = 3.38LADD60 pKa = 3.74EE61 pKa = 4.38VVTPLKK67 pKa = 8.49TTLSDD72 pKa = 3.5TLRR75 pKa = 11.84NPINQINDD83 pKa = 2.72IFRR86 pKa = 11.84IVALDD91 pKa = 3.39IPLQVTSIQKK101 pKa = 10.27DD102 pKa = 3.48LASQFSMLIDD112 pKa = 3.39SLNAIKK118 pKa = 10.61LGNGTNLIIPTSDD131 pKa = 2.77KK132 pKa = 10.71EE133 pKa = 4.18YY134 pKa = 10.99AGGIGNPVFTVDD146 pKa = 3.25AGGSIGFKK154 pKa = 9.57QFSLIEE160 pKa = 4.29HH161 pKa = 7.17PSFIAGPTTTRR172 pKa = 11.84GCTRR176 pKa = 11.84IPTFHH181 pKa = 6.76MSEE184 pKa = 4.21SHH186 pKa = 5.05WCYY189 pKa = 9.94SHH191 pKa = 7.04NIIAAGCQDD200 pKa = 3.49ASASSMYY207 pKa = 10.46ISMGVLHH214 pKa = 6.54VSSSGTPIFLTTASEE229 pKa = 4.59LIDD232 pKa = 4.71DD233 pKa = 4.47GVNRR237 pKa = 11.84KK238 pKa = 9.16SCSIVATQFGCDD250 pKa = 3.31ILCSIVIEE258 pKa = 4.95KK259 pKa = 10.73EE260 pKa = 3.82GDD262 pKa = 3.87DD263 pKa = 3.97YY264 pKa = 11.52WSDD267 pKa = 3.24TPTPMRR273 pKa = 11.84HH274 pKa = 5.49GRR276 pKa = 11.84FSFNGSFVEE285 pKa = 4.39TEE287 pKa = 4.37LPVSSMFSSFSANYY301 pKa = 8.0PAVGSGEE308 pKa = 4.07IVKK311 pKa = 10.47DD312 pKa = 3.88RR313 pKa = 11.84ILFPIYY319 pKa = 10.55GGIKK323 pKa = 7.85QTSPEE328 pKa = 3.75FTEE331 pKa = 4.16LVKK334 pKa = 11.22YY335 pKa = 10.5GLFVSTPTTVCQSSWTYY352 pKa = 11.11DD353 pKa = 3.39QVKK356 pKa = 9.54AAYY359 pKa = 9.45RR360 pKa = 11.84PDD362 pKa = 4.15YY363 pKa = 10.62ISGRR367 pKa = 11.84FWAQVILSCALDD379 pKa = 3.78AVDD382 pKa = 5.94LSSCIVKK389 pKa = 10.13IMNSSTVMMAAEE401 pKa = 4.06GRR403 pKa = 11.84IIKK406 pKa = 10.18IGIDD410 pKa = 3.03YY411 pKa = 9.78FYY413 pKa = 10.53YY414 pKa = 10.3QRR416 pKa = 11.84SSSWWPLAFVTKK428 pKa = 10.57LDD430 pKa = 3.92PQEE433 pKa = 4.79LADD436 pKa = 4.56TNSIWLTNSIPIPQSKK452 pKa = 9.59FPRR455 pKa = 11.84PSYY458 pKa = 10.67SEE460 pKa = 4.17NYY462 pKa = 6.58CTKK465 pKa = 10.29PAVCPATCVTGVYY478 pKa = 10.43SDD480 pKa = 3.44IWPLTSSSSLPSIIWIGQYY499 pKa = 10.79LDD501 pKa = 3.82APVGRR506 pKa = 11.84TYY508 pKa = 11.06PRR510 pKa = 11.84FGIANQSHH518 pKa = 6.99WYY520 pKa = 8.34LQEE523 pKa = 5.89DD524 pKa = 3.92ILPTSTASAYY534 pKa = 8.49STTTCFKK541 pKa = 8.14NTARR545 pKa = 11.84NRR547 pKa = 11.84VFCVTIAEE555 pKa = 4.31FADD558 pKa = 3.97GLFGEE563 pKa = 4.55YY564 pKa = 9.99RR565 pKa = 11.84ITPQLYY571 pKa = 9.5EE572 pKa = 3.99LVRR575 pKa = 11.84NNN577 pKa = 3.26
Molecular weight: 63.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C5I0V8|C5I0V8_9MONO RNA-directed RNA polymerase L OS=Avian metaavulavirus 8 OX=2560318 GN=L PE=3 SV=1
MM1 pKa = 8.04AYY3 pKa = 6.79TTLKK7 pKa = 10.58LWVDD11 pKa = 3.86EE12 pKa = 4.58GDD14 pKa = 3.52MSSSLLSFPLVLKK27 pKa = 9.21EE28 pKa = 4.05TDD30 pKa = 3.02RR31 pKa = 11.84GTKK34 pKa = 9.82KK35 pKa = 10.22LQPQVRR41 pKa = 11.84VDD43 pKa = 4.33SIGDD47 pKa = 3.38VQNAKK52 pKa = 9.96EE53 pKa = 3.97SSIFVTLYY61 pKa = 11.23GFIQAIKK68 pKa = 9.73EE69 pKa = 3.89NSDD72 pKa = 3.02RR73 pKa = 11.84SKK75 pKa = 10.9FFHH78 pKa = 6.92PKK80 pKa = 10.37DD81 pKa = 3.51DD82 pKa = 4.65FKK84 pKa = 11.56PEE86 pKa = 3.84TVTAGLVVVGAIRR99 pKa = 11.84MMADD103 pKa = 2.93VNTISNDD110 pKa = 3.06ALALEE115 pKa = 4.32ITVKK119 pKa = 10.55KK120 pKa = 10.59SATSQEE126 pKa = 3.74KK127 pKa = 6.8MTVMFHH133 pKa = 6.8NSPPSLRR140 pKa = 11.84TAITIRR146 pKa = 11.84AGGFISNADD155 pKa = 3.83EE156 pKa = 4.63NIKK159 pKa = 10.53CASKK163 pKa = 9.27LTAGVQYY170 pKa = 10.19IFRR173 pKa = 11.84PMFVSITKK181 pKa = 9.55LHH183 pKa = 5.95NGKK186 pKa = 10.22LYY188 pKa = 10.36RR189 pKa = 11.84VPKK192 pKa = 9.32SIHH195 pKa = 6.47SISSTLLYY203 pKa = 10.91SVMLEE208 pKa = 3.83VGFKK212 pKa = 10.35VDD214 pKa = 3.07IGKK217 pKa = 8.82DD218 pKa = 3.26HH219 pKa = 7.14PQAKK223 pKa = 8.32MLKK226 pKa = 9.34RR227 pKa = 11.84VTIGDD232 pKa = 3.87ADD234 pKa = 4.42TYY236 pKa = 10.93WGFAWFHH243 pKa = 6.12LCNFKK248 pKa = 9.49KK249 pKa = 9.27TSSKK253 pKa = 10.71GKK255 pKa = 9.35PRR257 pKa = 11.84TLDD260 pKa = 3.28EE261 pKa = 5.04LRR263 pKa = 11.84TKK265 pKa = 9.75VKK267 pKa = 10.91NMGLKK272 pKa = 10.58LEE274 pKa = 4.77LHH276 pKa = 6.76DD277 pKa = 4.35LWGPTIVVQITGKK290 pKa = 9.42SSKK293 pKa = 9.55YY294 pKa = 8.99AQGFFSSNGTCCLPISRR311 pKa = 11.84SAPEE315 pKa = 4.14LGKK318 pKa = 10.77LLWSCSATIGDD329 pKa = 4.04ATVVIQSSEE338 pKa = 3.84KK339 pKa = 11.01GEE341 pKa = 4.13LLRR344 pKa = 11.84SDD346 pKa = 3.95DD347 pKa = 3.93LEE349 pKa = 4.55IRR351 pKa = 11.84GAVASKK357 pKa = 10.37KK358 pKa = 10.03GRR360 pKa = 11.84LSSFHH365 pKa = 6.72PFKK368 pKa = 10.86KK369 pKa = 10.55
MM1 pKa = 8.04AYY3 pKa = 6.79TTLKK7 pKa = 10.58LWVDD11 pKa = 3.86EE12 pKa = 4.58GDD14 pKa = 3.52MSSSLLSFPLVLKK27 pKa = 9.21EE28 pKa = 4.05TDD30 pKa = 3.02RR31 pKa = 11.84GTKK34 pKa = 9.82KK35 pKa = 10.22LQPQVRR41 pKa = 11.84VDD43 pKa = 4.33SIGDD47 pKa = 3.38VQNAKK52 pKa = 9.96EE53 pKa = 3.97SSIFVTLYY61 pKa = 11.23GFIQAIKK68 pKa = 9.73EE69 pKa = 3.89NSDD72 pKa = 3.02RR73 pKa = 11.84SKK75 pKa = 10.9FFHH78 pKa = 6.92PKK80 pKa = 10.37DD81 pKa = 3.51DD82 pKa = 4.65FKK84 pKa = 11.56PEE86 pKa = 3.84TVTAGLVVVGAIRR99 pKa = 11.84MMADD103 pKa = 2.93VNTISNDD110 pKa = 3.06ALALEE115 pKa = 4.32ITVKK119 pKa = 10.55KK120 pKa = 10.59SATSQEE126 pKa = 3.74KK127 pKa = 6.8MTVMFHH133 pKa = 6.8NSPPSLRR140 pKa = 11.84TAITIRR146 pKa = 11.84AGGFISNADD155 pKa = 3.83EE156 pKa = 4.63NIKK159 pKa = 10.53CASKK163 pKa = 9.27LTAGVQYY170 pKa = 10.19IFRR173 pKa = 11.84PMFVSITKK181 pKa = 9.55LHH183 pKa = 5.95NGKK186 pKa = 10.22LYY188 pKa = 10.36RR189 pKa = 11.84VPKK192 pKa = 9.32SIHH195 pKa = 6.47SISSTLLYY203 pKa = 10.91SVMLEE208 pKa = 3.83VGFKK212 pKa = 10.35VDD214 pKa = 3.07IGKK217 pKa = 8.82DD218 pKa = 3.26HH219 pKa = 7.14PQAKK223 pKa = 8.32MLKK226 pKa = 9.34RR227 pKa = 11.84VTIGDD232 pKa = 3.87ADD234 pKa = 4.42TYY236 pKa = 10.93WGFAWFHH243 pKa = 6.12LCNFKK248 pKa = 9.49KK249 pKa = 9.27TSSKK253 pKa = 10.71GKK255 pKa = 9.35PRR257 pKa = 11.84TLDD260 pKa = 3.28EE261 pKa = 5.04LRR263 pKa = 11.84TKK265 pKa = 9.75VKK267 pKa = 10.91NMGLKK272 pKa = 10.58LEE274 pKa = 4.77LHH276 pKa = 6.76DD277 pKa = 4.35LWGPTIVVQITGKK290 pKa = 9.42SSKK293 pKa = 9.55YY294 pKa = 8.99AQGFFSSNGTCCLPISRR311 pKa = 11.84SAPEE315 pKa = 4.14LGKK318 pKa = 10.77LLWSCSATIGDD329 pKa = 4.04ATVVIQSSEE338 pKa = 3.84KK339 pKa = 11.01GEE341 pKa = 4.13LLRR344 pKa = 11.84SDD346 pKa = 3.95DD347 pKa = 3.93LEE349 pKa = 4.55IRR351 pKa = 11.84GAVASKK357 pKa = 10.37KK358 pKa = 10.03GRR360 pKa = 11.84LSSFHH365 pKa = 6.72PFKK368 pKa = 10.86KK369 pKa = 10.55
Molecular weight: 40.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5003 |
172 |
2238 |
625.4 |
69.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.056 ± 0.869 | 1.939 ± 0.306 |
5.297 ± 0.311 | 5.277 ± 0.458 |
3.238 ± 0.55 | 5.177 ± 0.464 |
1.919 ± 0.414 | 7.635 ± 0.54 |
5.237 ± 0.548 | 10.194 ± 1.205 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.219 ± 0.27 | 5.017 ± 0.389 |
4.517 ± 0.591 | 4.657 ± 0.559 |
4.617 ± 0.354 | 10.014 ± 0.328 |
6.976 ± 0.547 | 5.956 ± 0.474 |
1.099 ± 0.185 | 2.958 ± 0.478 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
