
Cryobacterium levicorallinum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Cryobacterium
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
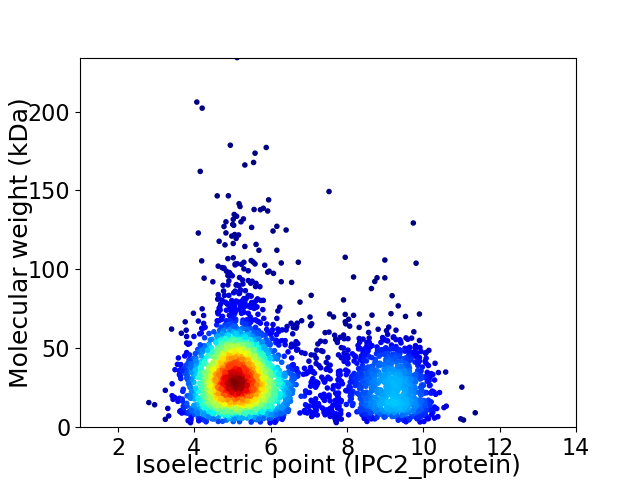
Virtual 2D-PAGE plot for 3509 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I2XXW3|A0A1I2XXW3_9MICO 50S ribosomal protein L34 OS=Cryobacterium levicorallinum OX=995038 GN=rpmH PE=3 SV=1
MM1 pKa = 7.47HH2 pKa = 7.23KK3 pKa = 10.1RR4 pKa = 11.84YY5 pKa = 8.96FAVPVLLASAALALSGCSAGTTASADD31 pKa = 3.69SADD34 pKa = 3.61VGLKK38 pKa = 10.02VVATTTQVADD48 pKa = 4.33FARR51 pKa = 11.84NVIGDD56 pKa = 3.71ASGVDD61 pKa = 4.26LTQLIQPNQSAHH73 pKa = 6.64SYY75 pKa = 10.52DD76 pKa = 3.94PSAADD81 pKa = 3.61LTALGAADD89 pKa = 3.34VLVINGVGLEE99 pKa = 4.03EE100 pKa = 4.29WLDD103 pKa = 3.76DD104 pKa = 4.98AIVASGFDD112 pKa = 3.82GVTIDD117 pKa = 3.93ADD119 pKa = 3.64EE120 pKa = 5.81GITISEE126 pKa = 4.1VGAGEE131 pKa = 4.08DD132 pKa = 3.52HH133 pKa = 6.75SDD135 pKa = 3.44EE136 pKa = 4.2VAEE139 pKa = 4.63EE140 pKa = 4.37DD141 pKa = 3.93DD142 pKa = 4.65HH143 pKa = 9.79ADD145 pKa = 3.72EE146 pKa = 5.12EE147 pKa = 5.18ADD149 pKa = 3.32AHH151 pKa = 6.79AGEE154 pKa = 5.55AEE156 pKa = 4.12DD157 pKa = 4.36AHH159 pKa = 7.9DD160 pKa = 5.34GGDD163 pKa = 3.41PHH165 pKa = 8.36IWTDD169 pKa = 2.8VHH171 pKa = 5.95NAEE174 pKa = 5.15TIVQTITDD182 pKa = 4.14GLVAADD188 pKa = 4.45SDD190 pKa = 3.78NAANFEE196 pKa = 4.52ANAAAYY202 pKa = 9.01SAQLSALDD210 pKa = 3.34DD211 pKa = 4.6WIRR214 pKa = 11.84TNVEE218 pKa = 4.11TVPEE222 pKa = 4.09SDD224 pKa = 3.56RR225 pKa = 11.84LLVSNHH231 pKa = 6.65DD232 pKa = 3.54ALGYY236 pKa = 7.72FTAAYY241 pKa = 9.53HH242 pKa = 6.12IDD244 pKa = 3.77YY245 pKa = 9.54VGSVIPSFDD254 pKa = 4.23DD255 pKa = 3.41NAEE258 pKa = 4.03PSAAEE263 pKa = 3.85IDD265 pKa = 4.03SLVAAITATGVTAVFSEE282 pKa = 4.56ASLSPKK288 pKa = 8.61TAEE291 pKa = 4.44TIATEE296 pKa = 4.17AGVTVYY302 pKa = 10.77SGDD305 pKa = 3.44DD306 pKa = 3.27ALYY309 pKa = 10.77VDD311 pKa = 4.6SLGPAGSAGDD321 pKa = 3.93TYY323 pKa = 11.22IKK325 pKa = 10.66AQLHH329 pKa = 6.16NSTLMLEE336 pKa = 4.39SWGVTPSALPAEE348 pKa = 4.55LQQ350 pKa = 3.33
MM1 pKa = 7.47HH2 pKa = 7.23KK3 pKa = 10.1RR4 pKa = 11.84YY5 pKa = 8.96FAVPVLLASAALALSGCSAGTTASADD31 pKa = 3.69SADD34 pKa = 3.61VGLKK38 pKa = 10.02VVATTTQVADD48 pKa = 4.33FARR51 pKa = 11.84NVIGDD56 pKa = 3.71ASGVDD61 pKa = 4.26LTQLIQPNQSAHH73 pKa = 6.64SYY75 pKa = 10.52DD76 pKa = 3.94PSAADD81 pKa = 3.61LTALGAADD89 pKa = 3.34VLVINGVGLEE99 pKa = 4.03EE100 pKa = 4.29WLDD103 pKa = 3.76DD104 pKa = 4.98AIVASGFDD112 pKa = 3.82GVTIDD117 pKa = 3.93ADD119 pKa = 3.64EE120 pKa = 5.81GITISEE126 pKa = 4.1VGAGEE131 pKa = 4.08DD132 pKa = 3.52HH133 pKa = 6.75SDD135 pKa = 3.44EE136 pKa = 4.2VAEE139 pKa = 4.63EE140 pKa = 4.37DD141 pKa = 3.93DD142 pKa = 4.65HH143 pKa = 9.79ADD145 pKa = 3.72EE146 pKa = 5.12EE147 pKa = 5.18ADD149 pKa = 3.32AHH151 pKa = 6.79AGEE154 pKa = 5.55AEE156 pKa = 4.12DD157 pKa = 4.36AHH159 pKa = 7.9DD160 pKa = 5.34GGDD163 pKa = 3.41PHH165 pKa = 8.36IWTDD169 pKa = 2.8VHH171 pKa = 5.95NAEE174 pKa = 5.15TIVQTITDD182 pKa = 4.14GLVAADD188 pKa = 4.45SDD190 pKa = 3.78NAANFEE196 pKa = 4.52ANAAAYY202 pKa = 9.01SAQLSALDD210 pKa = 3.34DD211 pKa = 4.6WIRR214 pKa = 11.84TNVEE218 pKa = 4.11TVPEE222 pKa = 4.09SDD224 pKa = 3.56RR225 pKa = 11.84LLVSNHH231 pKa = 6.65DD232 pKa = 3.54ALGYY236 pKa = 7.72FTAAYY241 pKa = 9.53HH242 pKa = 6.12IDD244 pKa = 3.77YY245 pKa = 9.54VGSVIPSFDD254 pKa = 4.23DD255 pKa = 3.41NAEE258 pKa = 4.03PSAAEE263 pKa = 3.85IDD265 pKa = 4.03SLVAAITATGVTAVFSEE282 pKa = 4.56ASLSPKK288 pKa = 8.61TAEE291 pKa = 4.44TIATEE296 pKa = 4.17AGVTVYY302 pKa = 10.77SGDD305 pKa = 3.44DD306 pKa = 3.27ALYY309 pKa = 10.77VDD311 pKa = 4.6SLGPAGSAGDD321 pKa = 3.93TYY323 pKa = 11.22IKK325 pKa = 10.66AQLHH329 pKa = 6.16NSTLMLEE336 pKa = 4.39SWGVTPSALPAEE348 pKa = 4.55LQQ350 pKa = 3.33
Molecular weight: 35.98 kDa
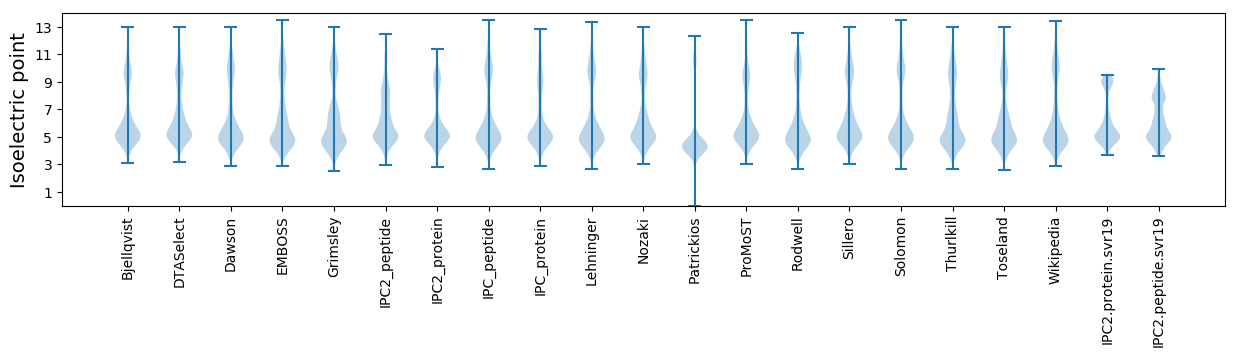
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3DHF3|A0A1I3DHF3_9MICO SipW-cognate class signal peptide OS=Cryobacterium levicorallinum OX=995038 GN=E3O11_09355 PE=4 SV=1
MM1 pKa = 7.76SIGILLVLAGLYY13 pKa = 10.12LLLMRR18 pKa = 11.84RR19 pKa = 11.84FVLRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84IAQGLPVSGPWHH38 pKa = 5.88FPAWLRR44 pKa = 11.84PRR46 pKa = 11.84VPRR49 pKa = 11.84WARR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84FARR57 pKa = 11.84RR58 pKa = 11.84PRR60 pKa = 11.84RR61 pKa = 11.84PRR63 pKa = 11.84AARR66 pKa = 11.84RR67 pKa = 11.84LRR69 pKa = 11.84HH70 pKa = 5.57TRR72 pKa = 11.84PGPPP76 pKa = 3.11
MM1 pKa = 7.76SIGILLVLAGLYY13 pKa = 10.12LLLMRR18 pKa = 11.84RR19 pKa = 11.84FVLRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84IAQGLPVSGPWHH38 pKa = 5.88FPAWLRR44 pKa = 11.84PRR46 pKa = 11.84VPRR49 pKa = 11.84WARR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84FARR57 pKa = 11.84RR58 pKa = 11.84PRR60 pKa = 11.84RR61 pKa = 11.84PRR63 pKa = 11.84AARR66 pKa = 11.84RR67 pKa = 11.84LRR69 pKa = 11.84HH70 pKa = 5.57TRR72 pKa = 11.84PGPPP76 pKa = 3.11
Molecular weight: 9.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1112413 |
25 |
2149 |
317.0 |
33.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.854 ± 0.053 | 0.582 ± 0.01 |
5.798 ± 0.036 | 5.163 ± 0.036 |
3.297 ± 0.027 | 8.634 ± 0.036 |
1.996 ± 0.02 | 5.033 ± 0.035 |
2.315 ± 0.025 | 10.548 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.864 ± 0.017 | 2.508 ± 0.025 |
5.149 ± 0.027 | 3.008 ± 0.022 |
6.56 ± 0.041 | 6.163 ± 0.029 |
6.447 ± 0.033 | 8.66 ± 0.034 |
1.394 ± 0.017 | 2.026 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
