
Cystobacter fuscus DSM 2262
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Cystobacterineae; Archangiaceae; Cystobacter; Cystobacter fuscus
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
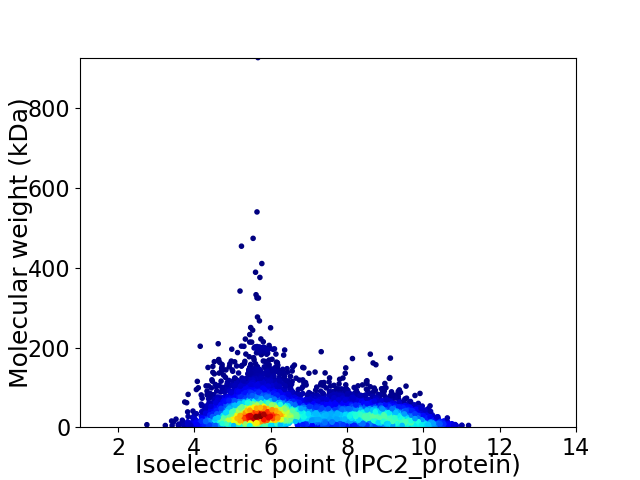
Virtual 2D-PAGE plot for 10511 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
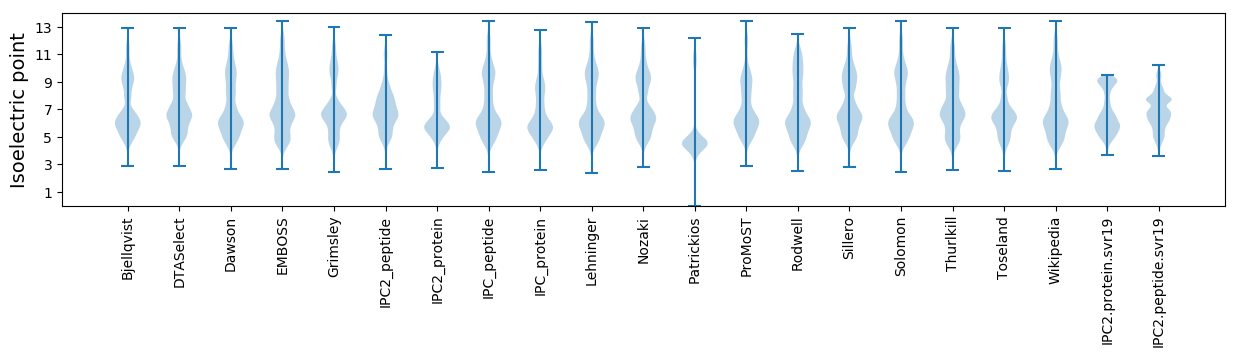
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S9QTE0|S9QTE0_9DELT Uncharacterized protein OS=Cystobacter fuscus DSM 2262 OX=1242864 GN=D187_002672 PE=4 SV=1
MM1 pKa = 7.15GWALALALMGAQLGCGSDD19 pKa = 3.42SDD21 pKa = 4.58GDD23 pKa = 4.42GIHH26 pKa = 7.54DD27 pKa = 4.79GADD30 pKa = 3.43CAPDD34 pKa = 3.97SDD36 pKa = 5.19SSWKK40 pKa = 9.73QLTMYY45 pKa = 10.95DD46 pKa = 3.98DD47 pKa = 4.89ADD49 pKa = 3.65GDD51 pKa = 4.17KK52 pKa = 10.84YY53 pKa = 11.37GSGEE57 pKa = 3.85PRR59 pKa = 11.84MICIGKK65 pKa = 7.26TLPPGLSTQGGDD77 pKa = 4.32CAPEE81 pKa = 4.27DD82 pKa = 3.54ATRR85 pKa = 11.84WMSLTGKK92 pKa = 10.25GYY94 pKa = 10.68YY95 pKa = 10.22ADD97 pKa = 4.42ADD99 pKa = 3.95GDD101 pKa = 4.1GKK103 pKa = 10.77AVHH106 pKa = 6.75APSSDD111 pKa = 3.35CVGANTTGYY120 pKa = 10.77LRR122 pKa = 11.84VQGTDD127 pKa = 4.11CDD129 pKa = 4.18DD130 pKa = 4.74ADD132 pKa = 3.88PKK134 pKa = 11.09VWTSRR139 pKa = 11.84SLYY142 pKa = 10.58ADD144 pKa = 3.48EE145 pKa = 6.35DD146 pKa = 4.51GDD148 pKa = 4.23GVAEE152 pKa = 4.63GEE154 pKa = 4.47ASLHH158 pKa = 6.49CIGSSVPPGMSEE170 pKa = 4.06TASDD174 pKa = 4.31CAPGDD179 pKa = 3.63STRR182 pKa = 11.84YY183 pKa = 7.96RR184 pKa = 11.84TWNHH188 pKa = 5.15VGRR191 pKa = 11.84DD192 pKa = 3.37EE193 pKa = 5.73DD194 pKa = 4.16GDD196 pKa = 4.16GFFVSAPGTLCIGEE210 pKa = 4.7APPAGYY216 pKa = 8.67TSSTLTRR223 pKa = 11.84LDD225 pKa = 4.03CDD227 pKa = 3.63DD228 pKa = 3.71TRR230 pKa = 11.84ADD232 pKa = 2.83VWTPLVQYY240 pKa = 10.42LDD242 pKa = 3.6SDD244 pKa = 4.05GDD246 pKa = 3.94QVGSGPAEE254 pKa = 4.06TLCSGEE260 pKa = 4.09GVRR263 pKa = 11.84PGYY266 pKa = 10.55SLGAGDD272 pKa = 5.64CAPDD276 pKa = 4.62DD277 pKa = 3.7NTRR280 pKa = 11.84WMLHH284 pKa = 5.38SYY286 pKa = 9.81LWRR289 pKa = 11.84DD290 pKa = 2.96ADD292 pKa = 3.84GDD294 pKa = 4.19GATVSQTGQVCGGFQIPAGFSMTPGTHH321 pKa = 7.05LDD323 pKa = 3.75CDD325 pKa = 3.71DD326 pKa = 3.55TRR328 pKa = 11.84AEE330 pKa = 4.32VSVTWSVFPDD340 pKa = 3.42ADD342 pKa = 3.47QDD344 pKa = 4.17GVGAGAQRR352 pKa = 11.84TICAGTTRR360 pKa = 11.84PAGYY364 pKa = 10.4ADD366 pKa = 4.19TGTDD370 pKa = 3.87CAPDD374 pKa = 3.62DD375 pKa = 3.92ALRR378 pKa = 11.84WRR380 pKa = 11.84TFTPRR385 pKa = 11.84FRR387 pKa = 11.84DD388 pKa = 3.25TDD390 pKa = 3.51GDD392 pKa = 4.21GFTVAPGGPVCLGASTQGYY411 pKa = 8.43PDD413 pKa = 3.47TSRR416 pKa = 11.84GTDD419 pKa = 3.33CDD421 pKa = 3.53DD422 pKa = 3.4TRR424 pKa = 11.84AEE426 pKa = 4.67VYY428 pKa = 10.3QSLQVHH434 pKa = 5.98VDD436 pKa = 3.09MDD438 pKa = 4.32GDD440 pKa = 4.51GVGSGPALTLCTNGSPPAGHH460 pKa = 6.58SLQGTDD466 pKa = 4.64CAPDD470 pKa = 3.71DD471 pKa = 3.92ASRR474 pKa = 11.84WRR476 pKa = 11.84LHH478 pKa = 5.29AFKK481 pKa = 10.92YY482 pKa = 10.6VDD484 pKa = 3.79ADD486 pKa = 3.87GDD488 pKa = 4.14GHH490 pKa = 5.95TLPSTGQVCAAATLPPPYY508 pKa = 9.68TPSASGNDD516 pKa = 4.03CDD518 pKa = 5.47DD519 pKa = 4.51ADD521 pKa = 3.88PARR524 pKa = 11.84FLWRR528 pKa = 11.84VLYY531 pKa = 9.75PDD533 pKa = 4.0RR534 pKa = 11.84DD535 pKa = 3.57GDD537 pKa = 4.54GVGTPPRR544 pKa = 11.84VVLCLDD550 pKa = 4.47DD551 pKa = 5.49APPPPGYY558 pKa = 10.05SIYY561 pKa = 10.97GFDD564 pKa = 5.41PDD566 pKa = 5.36DD567 pKa = 4.65SDD569 pKa = 4.0PASRR573 pKa = 11.84DD574 pKa = 3.36PADD577 pKa = 3.69EE578 pKa = 4.64SEE580 pKa = 4.24LDD582 pKa = 3.54EE583 pKa = 5.97FLFTQQ588 pKa = 4.37
MM1 pKa = 7.15GWALALALMGAQLGCGSDD19 pKa = 3.42SDD21 pKa = 4.58GDD23 pKa = 4.42GIHH26 pKa = 7.54DD27 pKa = 4.79GADD30 pKa = 3.43CAPDD34 pKa = 3.97SDD36 pKa = 5.19SSWKK40 pKa = 9.73QLTMYY45 pKa = 10.95DD46 pKa = 3.98DD47 pKa = 4.89ADD49 pKa = 3.65GDD51 pKa = 4.17KK52 pKa = 10.84YY53 pKa = 11.37GSGEE57 pKa = 3.85PRR59 pKa = 11.84MICIGKK65 pKa = 7.26TLPPGLSTQGGDD77 pKa = 4.32CAPEE81 pKa = 4.27DD82 pKa = 3.54ATRR85 pKa = 11.84WMSLTGKK92 pKa = 10.25GYY94 pKa = 10.68YY95 pKa = 10.22ADD97 pKa = 4.42ADD99 pKa = 3.95GDD101 pKa = 4.1GKK103 pKa = 10.77AVHH106 pKa = 6.75APSSDD111 pKa = 3.35CVGANTTGYY120 pKa = 10.77LRR122 pKa = 11.84VQGTDD127 pKa = 4.11CDD129 pKa = 4.18DD130 pKa = 4.74ADD132 pKa = 3.88PKK134 pKa = 11.09VWTSRR139 pKa = 11.84SLYY142 pKa = 10.58ADD144 pKa = 3.48EE145 pKa = 6.35DD146 pKa = 4.51GDD148 pKa = 4.23GVAEE152 pKa = 4.63GEE154 pKa = 4.47ASLHH158 pKa = 6.49CIGSSVPPGMSEE170 pKa = 4.06TASDD174 pKa = 4.31CAPGDD179 pKa = 3.63STRR182 pKa = 11.84YY183 pKa = 7.96RR184 pKa = 11.84TWNHH188 pKa = 5.15VGRR191 pKa = 11.84DD192 pKa = 3.37EE193 pKa = 5.73DD194 pKa = 4.16GDD196 pKa = 4.16GFFVSAPGTLCIGEE210 pKa = 4.7APPAGYY216 pKa = 8.67TSSTLTRR223 pKa = 11.84LDD225 pKa = 4.03CDD227 pKa = 3.63DD228 pKa = 3.71TRR230 pKa = 11.84ADD232 pKa = 2.83VWTPLVQYY240 pKa = 10.42LDD242 pKa = 3.6SDD244 pKa = 4.05GDD246 pKa = 3.94QVGSGPAEE254 pKa = 4.06TLCSGEE260 pKa = 4.09GVRR263 pKa = 11.84PGYY266 pKa = 10.55SLGAGDD272 pKa = 5.64CAPDD276 pKa = 4.62DD277 pKa = 3.7NTRR280 pKa = 11.84WMLHH284 pKa = 5.38SYY286 pKa = 9.81LWRR289 pKa = 11.84DD290 pKa = 2.96ADD292 pKa = 3.84GDD294 pKa = 4.19GATVSQTGQVCGGFQIPAGFSMTPGTHH321 pKa = 7.05LDD323 pKa = 3.75CDD325 pKa = 3.71DD326 pKa = 3.55TRR328 pKa = 11.84AEE330 pKa = 4.32VSVTWSVFPDD340 pKa = 3.42ADD342 pKa = 3.47QDD344 pKa = 4.17GVGAGAQRR352 pKa = 11.84TICAGTTRR360 pKa = 11.84PAGYY364 pKa = 10.4ADD366 pKa = 4.19TGTDD370 pKa = 3.87CAPDD374 pKa = 3.62DD375 pKa = 3.92ALRR378 pKa = 11.84WRR380 pKa = 11.84TFTPRR385 pKa = 11.84FRR387 pKa = 11.84DD388 pKa = 3.25TDD390 pKa = 3.51GDD392 pKa = 4.21GFTVAPGGPVCLGASTQGYY411 pKa = 8.43PDD413 pKa = 3.47TSRR416 pKa = 11.84GTDD419 pKa = 3.33CDD421 pKa = 3.53DD422 pKa = 3.4TRR424 pKa = 11.84AEE426 pKa = 4.67VYY428 pKa = 10.3QSLQVHH434 pKa = 5.98VDD436 pKa = 3.09MDD438 pKa = 4.32GDD440 pKa = 4.51GVGSGPALTLCTNGSPPAGHH460 pKa = 6.58SLQGTDD466 pKa = 4.64CAPDD470 pKa = 3.71DD471 pKa = 3.92ASRR474 pKa = 11.84WRR476 pKa = 11.84LHH478 pKa = 5.29AFKK481 pKa = 10.92YY482 pKa = 10.6VDD484 pKa = 3.79ADD486 pKa = 3.87GDD488 pKa = 4.14GHH490 pKa = 5.95TLPSTGQVCAAATLPPPYY508 pKa = 9.68TPSASGNDD516 pKa = 4.03CDD518 pKa = 5.47DD519 pKa = 4.51ADD521 pKa = 3.88PARR524 pKa = 11.84FLWRR528 pKa = 11.84VLYY531 pKa = 9.75PDD533 pKa = 4.0RR534 pKa = 11.84DD535 pKa = 3.57GDD537 pKa = 4.54GVGTPPRR544 pKa = 11.84VVLCLDD550 pKa = 4.47DD551 pKa = 5.49APPPPGYY558 pKa = 10.05SIYY561 pKa = 10.97GFDD564 pKa = 5.41PDD566 pKa = 5.36DD567 pKa = 4.65SDD569 pKa = 4.0PASRR573 pKa = 11.84DD574 pKa = 3.36PADD577 pKa = 3.69EE578 pKa = 4.64SEE580 pKa = 4.24LDD582 pKa = 3.54EE583 pKa = 5.97FLFTQQ588 pKa = 4.37
Molecular weight: 61.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S9PCR5|S9PCR5_9DELT ATEPR1 protein OS=Cystobacter fuscus DSM 2262 OX=1242864 GN=D187_001548 PE=4 SV=1
MM1 pKa = 7.85PSRR4 pKa = 11.84TRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84GGGWTWRR15 pKa = 11.84SAARR19 pKa = 11.84ARR21 pKa = 11.84GPVTHH26 pKa = 6.94ARR28 pKa = 11.84ASAPGALAPGGRR40 pKa = 11.84RR41 pKa = 11.84AA42 pKa = 3.86
MM1 pKa = 7.85PSRR4 pKa = 11.84TRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84GGGWTWRR15 pKa = 11.84SAARR19 pKa = 11.84ARR21 pKa = 11.84GPVTHH26 pKa = 6.94ARR28 pKa = 11.84ASAPGALAPGGRR40 pKa = 11.84RR41 pKa = 11.84AA42 pKa = 3.86
Molecular weight: 4.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3636354 |
37 |
8515 |
346.0 |
37.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.331 ± 0.034 | 0.948 ± 0.009 |
4.861 ± 0.015 | 6.632 ± 0.029 |
3.361 ± 0.015 | 8.714 ± 0.025 |
2.253 ± 0.012 | 3.054 ± 0.019 |
2.816 ± 0.022 | 11.228 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.962 ± 0.01 | 2.273 ± 0.019 |
6.111 ± 0.026 | 3.5 ± 0.015 |
8.098 ± 0.029 | 5.963 ± 0.021 |
5.416 ± 0.022 | 7.793 ± 0.019 |
1.487 ± 0.01 | 2.201 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
