
Bos taurus papillomavirus 19
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyoxipapillomavirus; Dyoxipapillomavirus 2
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
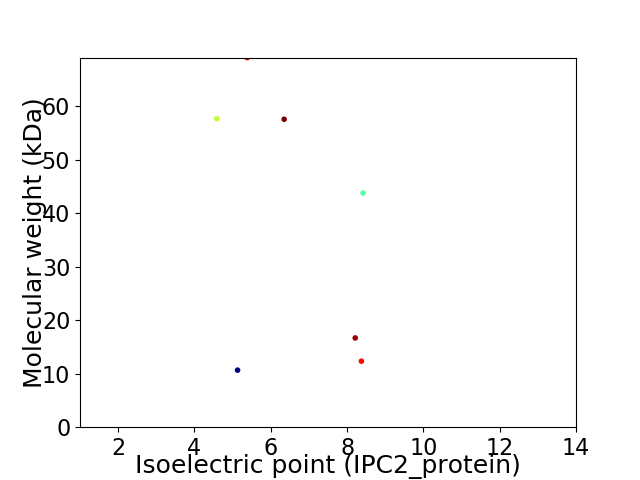
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
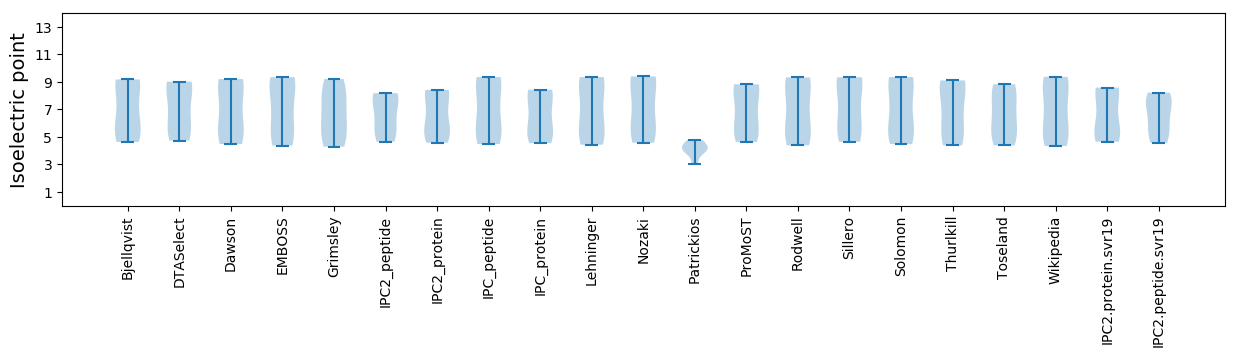
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B2K234|A0A1B2K234_9PAPI Regulatory protein E2 OS=Bos taurus papillomavirus 19 OX=1887217 GN=E2 PE=3 SV=1
MM1 pKa = 7.08TLRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.59RR8 pKa = 11.84DD9 pKa = 3.18SAEE12 pKa = 3.83NLWRR16 pKa = 11.84HH17 pKa = 5.31CQATGAEE24 pKa = 4.51CPPDD28 pKa = 4.03VINKK32 pKa = 6.84YY33 pKa = 8.0TEE35 pKa = 3.6NTLADD40 pKa = 3.99RR41 pKa = 11.84LSKK44 pKa = 10.5IFASILYY51 pKa = 10.15LGGLAIGTGKK61 pKa = 10.87GSGGNLGYY69 pKa = 10.38RR70 pKa = 11.84PLGEE74 pKa = 3.98PVGPPRR80 pKa = 11.84VGPGGTVIRR89 pKa = 11.84PNVVVDD95 pKa = 3.85AVGPADD101 pKa = 6.23LIPLDD106 pKa = 4.13SLNPDD111 pKa = 3.24SSVIPLLRR119 pKa = 11.84GTPEE123 pKa = 3.8GSEE126 pKa = 4.23VGLEE130 pKa = 4.14TPDD133 pKa = 3.36ILGEE137 pKa = 4.15SDD139 pKa = 3.49PASDD143 pKa = 3.79VTITTGNTVTLGEE156 pKa = 4.39DD157 pKa = 3.35TPAILEE163 pKa = 4.23VTPVQPEE170 pKa = 3.97EE171 pKa = 4.36AVPPPAKK178 pKa = 9.9RR179 pKa = 11.84PRR181 pKa = 11.84VSAQEE186 pKa = 4.09FLNATYY192 pKa = 10.46DD193 pKa = 3.5PSIFSTEE200 pKa = 3.96PVTLSTTGTASNSVTVDD217 pKa = 3.97FGLPSTEE224 pKa = 3.39IGQVFEE230 pKa = 4.91EE231 pKa = 4.42IEE233 pKa = 4.12LDD235 pKa = 3.37EE236 pKa = 4.37FTPLTRR242 pKa = 11.84SPQASTPADD251 pKa = 3.32EE252 pKa = 5.03SYY254 pKa = 11.79GLFSRR259 pKa = 11.84IRR261 pKa = 11.84EE262 pKa = 4.09FYY264 pKa = 10.03NRR266 pKa = 11.84RR267 pKa = 11.84IRR269 pKa = 11.84QVEE272 pKa = 3.97ITSPTFLRR280 pKa = 11.84RR281 pKa = 11.84PQAYY285 pKa = 8.51IDD287 pKa = 3.55FAYY290 pKa = 10.29GVDD293 pKa = 4.18NPAFDD298 pKa = 5.06PDD300 pKa = 3.37VSRR303 pKa = 11.84EE304 pKa = 3.88FLEE307 pKa = 5.4DD308 pKa = 3.36LAEE311 pKa = 4.37LEE313 pKa = 4.53NVTAAPHH320 pKa = 6.83SDD322 pKa = 3.19FQDD325 pKa = 2.88IVRR328 pKa = 11.84LNRR331 pKa = 11.84QVFQEE336 pKa = 4.54GEE338 pKa = 4.09GGRR341 pKa = 11.84LRR343 pKa = 11.84VSRR346 pKa = 11.84IGNRR350 pKa = 11.84GTISTRR356 pKa = 11.84SGLLIGEE363 pKa = 4.19RR364 pKa = 11.84VHH366 pKa = 7.34FYY368 pKa = 11.06QDD370 pKa = 3.17LSSIQQSDD378 pKa = 4.48GIEE381 pKa = 3.97LAPLGSFTGEE391 pKa = 3.8ATVVTGTEE399 pKa = 3.34EE400 pKa = 4.17GAFVDD405 pKa = 4.33ASSSFDD411 pKa = 3.58YY412 pKa = 10.81EE413 pKa = 4.21GTGIEE418 pKa = 5.98DD419 pKa = 3.58VLLDD423 pKa = 3.58NYY425 pKa = 10.49EE426 pKa = 4.08EE427 pKa = 4.37SFEE430 pKa = 4.27RR431 pKa = 11.84SQLVIGTRR439 pKa = 11.84RR440 pKa = 11.84SSNIVPMDD448 pKa = 3.89FYY450 pKa = 10.81RR451 pKa = 11.84GEE453 pKa = 3.9IRR455 pKa = 11.84PFVQDD460 pKa = 3.37LTEE463 pKa = 3.84GWFYY467 pKa = 11.32GPPEE471 pKa = 4.42IGNDD475 pKa = 3.64EE476 pKa = 4.11NTPIYY481 pKa = 10.19IDD483 pKa = 3.78WSPLEE488 pKa = 4.35PGSRR492 pKa = 11.84GPDD495 pKa = 3.67LIINLYY501 pKa = 9.85GNDD504 pKa = 3.64YY505 pKa = 10.49SLHH508 pKa = 6.19PSYY511 pKa = 11.27LLRR514 pKa = 11.84RR515 pKa = 11.84RR516 pKa = 11.84RR517 pKa = 11.84KK518 pKa = 8.33RR519 pKa = 11.84KK520 pKa = 9.47RR521 pKa = 11.84FHH523 pKa = 6.41VLL525 pKa = 2.81
MM1 pKa = 7.08TLRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.59RR8 pKa = 11.84DD9 pKa = 3.18SAEE12 pKa = 3.83NLWRR16 pKa = 11.84HH17 pKa = 5.31CQATGAEE24 pKa = 4.51CPPDD28 pKa = 4.03VINKK32 pKa = 6.84YY33 pKa = 8.0TEE35 pKa = 3.6NTLADD40 pKa = 3.99RR41 pKa = 11.84LSKK44 pKa = 10.5IFASILYY51 pKa = 10.15LGGLAIGTGKK61 pKa = 10.87GSGGNLGYY69 pKa = 10.38RR70 pKa = 11.84PLGEE74 pKa = 3.98PVGPPRR80 pKa = 11.84VGPGGTVIRR89 pKa = 11.84PNVVVDD95 pKa = 3.85AVGPADD101 pKa = 6.23LIPLDD106 pKa = 4.13SLNPDD111 pKa = 3.24SSVIPLLRR119 pKa = 11.84GTPEE123 pKa = 3.8GSEE126 pKa = 4.23VGLEE130 pKa = 4.14TPDD133 pKa = 3.36ILGEE137 pKa = 4.15SDD139 pKa = 3.49PASDD143 pKa = 3.79VTITTGNTVTLGEE156 pKa = 4.39DD157 pKa = 3.35TPAILEE163 pKa = 4.23VTPVQPEE170 pKa = 3.97EE171 pKa = 4.36AVPPPAKK178 pKa = 9.9RR179 pKa = 11.84PRR181 pKa = 11.84VSAQEE186 pKa = 4.09FLNATYY192 pKa = 10.46DD193 pKa = 3.5PSIFSTEE200 pKa = 3.96PVTLSTTGTASNSVTVDD217 pKa = 3.97FGLPSTEE224 pKa = 3.39IGQVFEE230 pKa = 4.91EE231 pKa = 4.42IEE233 pKa = 4.12LDD235 pKa = 3.37EE236 pKa = 4.37FTPLTRR242 pKa = 11.84SPQASTPADD251 pKa = 3.32EE252 pKa = 5.03SYY254 pKa = 11.79GLFSRR259 pKa = 11.84IRR261 pKa = 11.84EE262 pKa = 4.09FYY264 pKa = 10.03NRR266 pKa = 11.84RR267 pKa = 11.84IRR269 pKa = 11.84QVEE272 pKa = 3.97ITSPTFLRR280 pKa = 11.84RR281 pKa = 11.84PQAYY285 pKa = 8.51IDD287 pKa = 3.55FAYY290 pKa = 10.29GVDD293 pKa = 4.18NPAFDD298 pKa = 5.06PDD300 pKa = 3.37VSRR303 pKa = 11.84EE304 pKa = 3.88FLEE307 pKa = 5.4DD308 pKa = 3.36LAEE311 pKa = 4.37LEE313 pKa = 4.53NVTAAPHH320 pKa = 6.83SDD322 pKa = 3.19FQDD325 pKa = 2.88IVRR328 pKa = 11.84LNRR331 pKa = 11.84QVFQEE336 pKa = 4.54GEE338 pKa = 4.09GGRR341 pKa = 11.84LRR343 pKa = 11.84VSRR346 pKa = 11.84IGNRR350 pKa = 11.84GTISTRR356 pKa = 11.84SGLLIGEE363 pKa = 4.19RR364 pKa = 11.84VHH366 pKa = 7.34FYY368 pKa = 11.06QDD370 pKa = 3.17LSSIQQSDD378 pKa = 4.48GIEE381 pKa = 3.97LAPLGSFTGEE391 pKa = 3.8ATVVTGTEE399 pKa = 3.34EE400 pKa = 4.17GAFVDD405 pKa = 4.33ASSSFDD411 pKa = 3.58YY412 pKa = 10.81EE413 pKa = 4.21GTGIEE418 pKa = 5.98DD419 pKa = 3.58VLLDD423 pKa = 3.58NYY425 pKa = 10.49EE426 pKa = 4.08EE427 pKa = 4.37SFEE430 pKa = 4.27RR431 pKa = 11.84SQLVIGTRR439 pKa = 11.84RR440 pKa = 11.84SSNIVPMDD448 pKa = 3.89FYY450 pKa = 10.81RR451 pKa = 11.84GEE453 pKa = 3.9IRR455 pKa = 11.84PFVQDD460 pKa = 3.37LTEE463 pKa = 3.84GWFYY467 pKa = 11.32GPPEE471 pKa = 4.42IGNDD475 pKa = 3.64EE476 pKa = 4.11NTPIYY481 pKa = 10.19IDD483 pKa = 3.78WSPLEE488 pKa = 4.35PGSRR492 pKa = 11.84GPDD495 pKa = 3.67LIINLYY501 pKa = 9.85GNDD504 pKa = 3.64YY505 pKa = 10.49SLHH508 pKa = 6.19PSYY511 pKa = 11.27LLRR514 pKa = 11.84RR515 pKa = 11.84RR516 pKa = 11.84RR517 pKa = 11.84KK518 pKa = 8.33RR519 pKa = 11.84KK520 pKa = 9.47RR521 pKa = 11.84FHH523 pKa = 6.41VLL525 pKa = 2.81
Molecular weight: 57.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B2K268|A0A1B2K268_9PAPI Replication protein E1 OS=Bos taurus papillomavirus 19 OX=1887217 GN=E1 PE=3 SV=1
MM1 pKa = 7.54KK2 pKa = 8.68PTVIVCFFMGKK13 pKa = 8.57EE14 pKa = 4.18SRR16 pKa = 11.84CMPGIFGPGMVRR28 pKa = 11.84LEE30 pKa = 3.77KK31 pKa = 10.34RR32 pKa = 11.84YY33 pKa = 9.77LQMEE37 pKa = 4.51KK38 pKa = 10.86LGMTFSLVPLVALAKK53 pKa = 10.43ALIIILLVPVGQWLPQMLTYY73 pKa = 9.9LTGHH77 pKa = 6.81FGCNEE82 pKa = 3.64HH83 pKa = 6.85KK84 pKa = 10.58AIIMALLGEE93 pKa = 4.49ISYY96 pKa = 10.61LLQWLIIPATQTILL110 pKa = 3.46
MM1 pKa = 7.54KK2 pKa = 8.68PTVIVCFFMGKK13 pKa = 8.57EE14 pKa = 4.18SRR16 pKa = 11.84CMPGIFGPGMVRR28 pKa = 11.84LEE30 pKa = 3.77KK31 pKa = 10.34RR32 pKa = 11.84YY33 pKa = 9.77LQMEE37 pKa = 4.51KK38 pKa = 10.86LGMTFSLVPLVALAKK53 pKa = 10.43ALIIILLVPVGQWLPQMLTYY73 pKa = 9.9LTGHH77 pKa = 6.81FGCNEE82 pKa = 3.64HH83 pKa = 6.85KK84 pKa = 10.58AIIMALLGEE93 pKa = 4.49ISYY96 pKa = 10.61LLQWLIIPATQTILL110 pKa = 3.46
Molecular weight: 12.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2383 |
95 |
612 |
340.4 |
38.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.581 ± 0.349 | 2.266 ± 0.696 |
6.085 ± 0.633 | 6.504 ± 0.635 |
4.532 ± 0.588 | 7.134 ± 0.873 |
1.888 ± 0.253 | 4.826 ± 0.615 |
5.455 ± 1.105 | 9.987 ± 0.964 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.804 ± 0.497 | 4.154 ± 0.565 |
5.581 ± 0.76 | 3.777 ± 0.306 |
6.085 ± 1.03 | 7.512 ± 0.761 |
6.211 ± 0.304 | 5.917 ± 0.361 |
1.553 ± 0.301 | 3.147 ± 0.325 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
