
Malpais Spring vesiculovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Vesiculovirus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
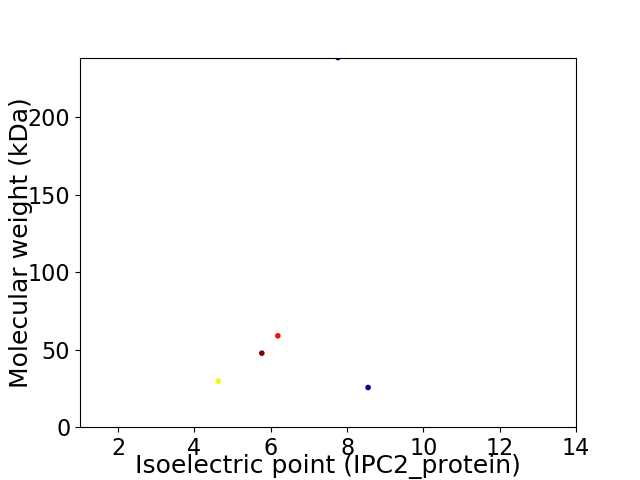
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
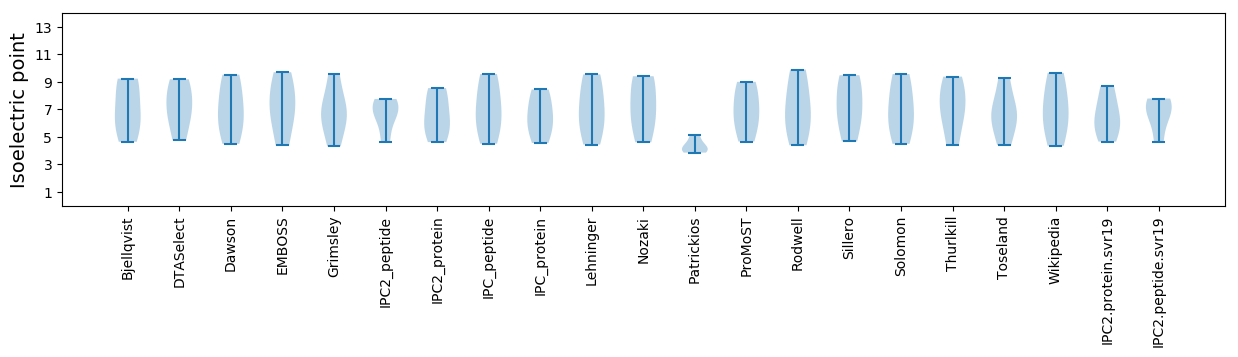
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M4VS08|M4VS08_9RHAB Matrix OS=Malpais Spring vesiculovirus OX=1972570 PE=3 SV=1
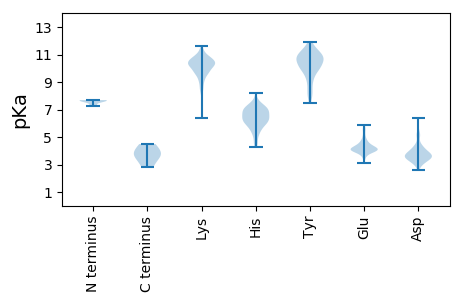
MM1 pKa = 7.27SKK3 pKa = 9.78IVEE6 pKa = 4.16LLKK9 pKa = 10.79NYY11 pKa = 9.17PNIMNTMEE19 pKa = 5.64DD20 pKa = 3.45IEE22 pKa = 4.42SLEE25 pKa = 4.46GEE27 pKa = 4.75VNPKK31 pKa = 9.67AASVVPGSSKK41 pKa = 10.89EE42 pKa = 4.1EE43 pKa = 3.85PTPSYY48 pKa = 10.71FLADD52 pKa = 3.63MLPEE56 pKa = 4.33EE57 pKa = 4.81EE58 pKa = 5.43GEE60 pKa = 4.16DD61 pKa = 3.4PKK63 pKa = 11.53GGVEE67 pKa = 3.84AGEE70 pKa = 4.6EE71 pKa = 4.14EE72 pKa = 5.07GSTAGDD78 pKa = 3.49YY79 pKa = 11.18DD80 pKa = 4.21DD81 pKa = 5.4YY82 pKa = 11.87AVQFEE87 pKa = 4.16DD88 pKa = 4.33RR89 pKa = 11.84EE90 pKa = 4.56WVAGTEE96 pKa = 3.99ISHH99 pKa = 7.45DD100 pKa = 3.77GTKK103 pKa = 10.08HH104 pKa = 4.27YY105 pKa = 11.14VITNPVKK112 pKa = 10.74SNRR115 pKa = 11.84EE116 pKa = 3.88LTKK119 pKa = 10.07KK120 pKa = 6.42WTAGIVGLLKK130 pKa = 10.49HH131 pKa = 6.84IEE133 pKa = 3.95EE134 pKa = 4.64GAGIKK139 pKa = 9.85INCEE143 pKa = 3.7EE144 pKa = 4.09VEE146 pKa = 4.16QGIKK150 pKa = 10.29CQMVTSCPPHH160 pKa = 6.93DD161 pKa = 5.22ANDD164 pKa = 3.93TSSSSGDD171 pKa = 3.61SEE173 pKa = 4.86TCSSPTSDD181 pKa = 3.27KK182 pKa = 11.21SKK184 pKa = 11.06NSTTISSVPDD194 pKa = 3.62FDD196 pKa = 4.36HH197 pKa = 7.32PSIIDD202 pKa = 4.4LIDD205 pKa = 4.32RR206 pKa = 11.84DD207 pKa = 3.71ITLPSLDD214 pKa = 3.68DD215 pKa = 3.81SKK217 pKa = 11.43PGFVLQLPRR226 pKa = 11.84LFGSRR231 pKa = 11.84EE232 pKa = 3.67AAINCCKK239 pKa = 10.36EE240 pKa = 4.01GEE242 pKa = 4.27VSIKK246 pKa = 10.22SAVVAGLRR254 pKa = 11.84RR255 pKa = 11.84KK256 pKa = 10.16GIYY259 pKa = 9.8NKK261 pKa = 9.68IRR263 pKa = 11.84IKK265 pKa = 10.76FDD267 pKa = 3.26LDD269 pKa = 3.65KK270 pKa = 11.25IQFF273 pKa = 3.63
MM1 pKa = 7.27SKK3 pKa = 9.78IVEE6 pKa = 4.16LLKK9 pKa = 10.79NYY11 pKa = 9.17PNIMNTMEE19 pKa = 5.64DD20 pKa = 3.45IEE22 pKa = 4.42SLEE25 pKa = 4.46GEE27 pKa = 4.75VNPKK31 pKa = 9.67AASVVPGSSKK41 pKa = 10.89EE42 pKa = 4.1EE43 pKa = 3.85PTPSYY48 pKa = 10.71FLADD52 pKa = 3.63MLPEE56 pKa = 4.33EE57 pKa = 4.81EE58 pKa = 5.43GEE60 pKa = 4.16DD61 pKa = 3.4PKK63 pKa = 11.53GGVEE67 pKa = 3.84AGEE70 pKa = 4.6EE71 pKa = 4.14EE72 pKa = 5.07GSTAGDD78 pKa = 3.49YY79 pKa = 11.18DD80 pKa = 4.21DD81 pKa = 5.4YY82 pKa = 11.87AVQFEE87 pKa = 4.16DD88 pKa = 4.33RR89 pKa = 11.84EE90 pKa = 4.56WVAGTEE96 pKa = 3.99ISHH99 pKa = 7.45DD100 pKa = 3.77GTKK103 pKa = 10.08HH104 pKa = 4.27YY105 pKa = 11.14VITNPVKK112 pKa = 10.74SNRR115 pKa = 11.84EE116 pKa = 3.88LTKK119 pKa = 10.07KK120 pKa = 6.42WTAGIVGLLKK130 pKa = 10.49HH131 pKa = 6.84IEE133 pKa = 3.95EE134 pKa = 4.64GAGIKK139 pKa = 9.85INCEE143 pKa = 3.7EE144 pKa = 4.09VEE146 pKa = 4.16QGIKK150 pKa = 10.29CQMVTSCPPHH160 pKa = 6.93DD161 pKa = 5.22ANDD164 pKa = 3.93TSSSSGDD171 pKa = 3.61SEE173 pKa = 4.86TCSSPTSDD181 pKa = 3.27KK182 pKa = 11.21SKK184 pKa = 11.06NSTTISSVPDD194 pKa = 3.62FDD196 pKa = 4.36HH197 pKa = 7.32PSIIDD202 pKa = 4.4LIDD205 pKa = 4.32RR206 pKa = 11.84DD207 pKa = 3.71ITLPSLDD214 pKa = 3.68DD215 pKa = 3.81SKK217 pKa = 11.43PGFVLQLPRR226 pKa = 11.84LFGSRR231 pKa = 11.84EE232 pKa = 3.67AAINCCKK239 pKa = 10.36EE240 pKa = 4.01GEE242 pKa = 4.27VSIKK246 pKa = 10.22SAVVAGLRR254 pKa = 11.84RR255 pKa = 11.84KK256 pKa = 10.16GIYY259 pKa = 9.8NKK261 pKa = 9.68IRR263 pKa = 11.84IKK265 pKa = 10.76FDD267 pKa = 3.26LDD269 pKa = 3.65KK270 pKa = 11.25IQFF273 pKa = 3.63
Molecular weight: 29.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M4W6E4|M4W6E4_9RHAB Nucleocapsid protein OS=Malpais Spring vesiculovirus OX=1972570 PE=3 SV=1
MM1 pKa = 7.59KK2 pKa = 10.25SIKK5 pKa = 9.98KK6 pKa = 10.04VLGKK10 pKa = 10.42KK11 pKa = 9.48KK12 pKa = 10.57EE13 pKa = 4.02KK14 pKa = 10.77GEE16 pKa = 4.23KK17 pKa = 9.68KK18 pKa = 9.9SKK20 pKa = 10.65KK21 pKa = 9.65YY22 pKa = 10.39DD23 pKa = 3.44LPPNYY28 pKa = 9.74NDD30 pKa = 5.62LIGPSAPSAPMFGLDD45 pKa = 3.16PSDD48 pKa = 3.59YY49 pKa = 10.9FEE51 pKa = 5.24QINSNDD57 pKa = 3.43SVVIKK62 pKa = 10.45LKK64 pKa = 10.54YY65 pKa = 8.32SCEE68 pKa = 4.05VQVRR72 pKa = 11.84AIRR75 pKa = 11.84PFSGVLEE82 pKa = 4.37AADD85 pKa = 5.2AIARR89 pKa = 11.84WEE91 pKa = 3.61MDD93 pKa = 2.96YY94 pKa = 11.35RR95 pKa = 11.84GFLGKK100 pKa = 10.36KK101 pKa = 8.36PFYY104 pKa = 9.88RR105 pKa = 11.84LLMGIAIKK113 pKa = 10.35KK114 pKa = 9.52LRR116 pKa = 11.84AAPSSLTEE124 pKa = 4.09GNRR127 pKa = 11.84PEE129 pKa = 4.15YY130 pKa = 9.98NCLFEE135 pKa = 3.96GHH137 pKa = 6.0GAIRR141 pKa = 11.84HH142 pKa = 5.61NLGQLPPMSYY152 pKa = 10.52VSEE155 pKa = 4.29TFTRR159 pKa = 11.84DD160 pKa = 2.58WQTDD164 pKa = 3.38KK165 pKa = 11.63NKK167 pKa = 10.64GSVHH171 pKa = 4.71VKK173 pKa = 9.73FWLGMLDD180 pKa = 3.92TNDD183 pKa = 3.91EE184 pKa = 4.18MPEE187 pKa = 3.91ILSSKK192 pKa = 9.89SFQSEE197 pKa = 4.49SEE199 pKa = 4.16LKK201 pKa = 10.63QIMEE205 pKa = 4.04MMGIRR210 pKa = 11.84VKK212 pKa = 10.61KK213 pKa = 10.77SKK215 pKa = 10.3DD216 pKa = 3.54NKK218 pKa = 9.51WEE220 pKa = 3.98ILSTCC225 pKa = 4.03
MM1 pKa = 7.59KK2 pKa = 10.25SIKK5 pKa = 9.98KK6 pKa = 10.04VLGKK10 pKa = 10.42KK11 pKa = 9.48KK12 pKa = 10.57EE13 pKa = 4.02KK14 pKa = 10.77GEE16 pKa = 4.23KK17 pKa = 9.68KK18 pKa = 9.9SKK20 pKa = 10.65KK21 pKa = 9.65YY22 pKa = 10.39DD23 pKa = 3.44LPPNYY28 pKa = 9.74NDD30 pKa = 5.62LIGPSAPSAPMFGLDD45 pKa = 3.16PSDD48 pKa = 3.59YY49 pKa = 10.9FEE51 pKa = 5.24QINSNDD57 pKa = 3.43SVVIKK62 pKa = 10.45LKK64 pKa = 10.54YY65 pKa = 8.32SCEE68 pKa = 4.05VQVRR72 pKa = 11.84AIRR75 pKa = 11.84PFSGVLEE82 pKa = 4.37AADD85 pKa = 5.2AIARR89 pKa = 11.84WEE91 pKa = 3.61MDD93 pKa = 2.96YY94 pKa = 11.35RR95 pKa = 11.84GFLGKK100 pKa = 10.36KK101 pKa = 8.36PFYY104 pKa = 9.88RR105 pKa = 11.84LLMGIAIKK113 pKa = 10.35KK114 pKa = 9.52LRR116 pKa = 11.84AAPSSLTEE124 pKa = 4.09GNRR127 pKa = 11.84PEE129 pKa = 4.15YY130 pKa = 9.98NCLFEE135 pKa = 3.96GHH137 pKa = 6.0GAIRR141 pKa = 11.84HH142 pKa = 5.61NLGQLPPMSYY152 pKa = 10.52VSEE155 pKa = 4.29TFTRR159 pKa = 11.84DD160 pKa = 2.58WQTDD164 pKa = 3.38KK165 pKa = 11.63NKK167 pKa = 10.64GSVHH171 pKa = 4.71VKK173 pKa = 9.73FWLGMLDD180 pKa = 3.92TNDD183 pKa = 3.91EE184 pKa = 4.18MPEE187 pKa = 3.91ILSSKK192 pKa = 9.89SFQSEE197 pKa = 4.49SEE199 pKa = 4.16LKK201 pKa = 10.63QIMEE205 pKa = 4.04MMGIRR210 pKa = 11.84VKK212 pKa = 10.61KK213 pKa = 10.77SKK215 pKa = 10.3DD216 pKa = 3.54NKK218 pKa = 9.51WEE220 pKa = 3.98ILSTCC225 pKa = 4.03
Molecular weight: 25.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3539 |
225 |
2095 |
707.8 |
80.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.662 ± 0.77 | 1.978 ± 0.304 |
6.019 ± 0.563 | 6.019 ± 0.668 |
3.956 ± 0.333 | 6.047 ± 0.486 |
2.515 ± 0.195 | 7.573 ± 0.331 |
7.262 ± 0.493 | 9.268 ± 0.992 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.458 ± 0.203 | 4.719 ± 0.615 |
5.199 ± 0.398 | 2.797 ± 0.14 |
4.832 ± 0.637 | 8.392 ± 0.475 |
5.736 ± 0.313 | 5.397 ± 0.419 |
1.893 ± 0.194 | 3.278 ± 0.259 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
