
Planococcus rifietoensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Planococcaceae; Planococcus
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
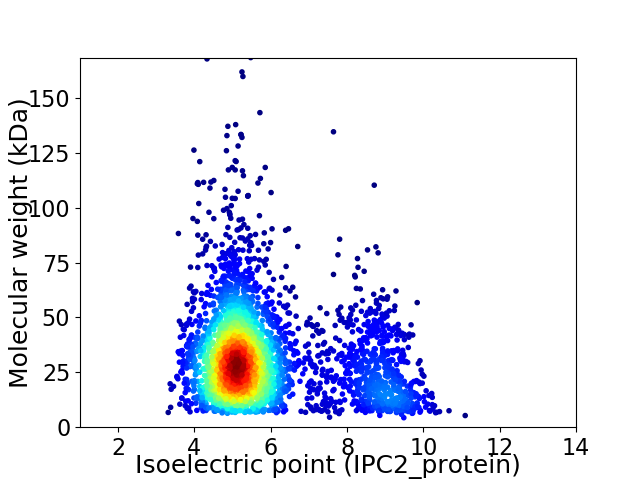
Virtual 2D-PAGE plot for 3386 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U2YV83|A0A0U2YV83_9BACL RNA methyltransferase TrmH OS=Planococcus rifietoensis OX=200991 GN=AUC31_09775 PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 10.37KK3 pKa = 10.32FNLMFLAAASMAALAACSSGDD24 pKa = 3.57EE25 pKa = 4.23EE26 pKa = 5.1ASGGSGSEE34 pKa = 4.07GSADD38 pKa = 3.41VDD40 pKa = 5.55EE41 pKa = 4.52ITAWAWDD48 pKa = 3.58PAFNIAALEE57 pKa = 4.13NAKK60 pKa = 9.89EE61 pKa = 4.09AYY63 pKa = 10.29NGDD66 pKa = 2.9GDD68 pKa = 4.27YY69 pKa = 11.04EE70 pKa = 4.56VNIIEE75 pKa = 4.5NAQDD79 pKa = 5.51DD80 pKa = 4.9IIQKK84 pKa = 10.54LNTGLSSGTTNGMPNIVLIEE104 pKa = 4.37DD105 pKa = 3.41YY106 pKa = 10.36RR107 pKa = 11.84AQSFLQAYY115 pKa = 8.23PDD117 pKa = 3.89AFHH120 pKa = 7.54PLTDD124 pKa = 4.61VINSDD129 pKa = 4.22DD130 pKa = 3.72FADD133 pKa = 3.78YY134 pKa = 10.8KK135 pKa = 10.78IATTSYY141 pKa = 10.91DD142 pKa = 3.31GEE144 pKa = 4.49QYY146 pKa = 11.35GLPFDD151 pKa = 4.24SGVAALYY158 pKa = 10.74VRR160 pKa = 11.84TDD162 pKa = 3.48YY163 pKa = 11.52LEE165 pKa = 4.05EE166 pKa = 5.61AGYY169 pKa = 10.44SVEE172 pKa = 4.85DD173 pKa = 3.52VTDD176 pKa = 3.66ITWQEE181 pKa = 3.99YY182 pKa = 10.34IEE184 pKa = 4.35IGKK187 pKa = 9.3DD188 pKa = 3.19VKK190 pKa = 10.77EE191 pKa = 4.02ATGKK195 pKa = 10.46NMLTLDD201 pKa = 4.5PNDD204 pKa = 3.71LAQVRR209 pKa = 11.84MMIQTNGSWYY219 pKa = 10.17VEE221 pKa = 4.27EE222 pKa = 5.93DD223 pKa = 3.24GSTPNIAGNEE233 pKa = 4.01TLEE236 pKa = 4.98AGFEE240 pKa = 4.5TYY242 pKa = 10.74KK243 pKa = 11.25EE244 pKa = 4.05MMDD247 pKa = 3.43ADD249 pKa = 3.42IAKK252 pKa = 9.81IVSDD256 pKa = 3.53WSQFVGAFNSGEE268 pKa = 4.24VASVPTGNWITPSVKK283 pKa = 9.92QAADD287 pKa = 3.49QSGKK291 pKa = 8.65WAVVPMPRR299 pKa = 11.84LDD301 pKa = 3.44MEE303 pKa = 4.69GAVNASNLGGSSWYY317 pKa = 9.91VLNVDD322 pKa = 4.65GKK324 pKa = 9.78EE325 pKa = 3.91QAADD329 pKa = 3.56FLLNTFGSNPEE340 pKa = 4.13LYY342 pKa = 8.53QTLVEE347 pKa = 5.37DD348 pKa = 3.86IGALGTYY355 pKa = 10.03SPAAEE360 pKa = 4.02GDD362 pKa = 3.47AYY364 pKa = 10.64AVEE367 pKa = 5.1DD368 pKa = 4.37EE369 pKa = 4.8FFGGQQLLTDD379 pKa = 4.45LSTWTDD385 pKa = 3.62EE386 pKa = 4.3IPAVNYY392 pKa = 10.0GLHH395 pKa = 6.22TYY397 pKa = 10.28AIEE400 pKa = 5.42DD401 pKa = 3.55ILVTGMQDD409 pKa = 3.54YY410 pKa = 11.43LNGAEE415 pKa = 4.93LDD417 pKa = 3.98TVLEE421 pKa = 4.19NVQGQAEE428 pKa = 4.28AQLKK432 pKa = 9.34
MM1 pKa = 7.56KK2 pKa = 10.37KK3 pKa = 10.32FNLMFLAAASMAALAACSSGDD24 pKa = 3.57EE25 pKa = 4.23EE26 pKa = 5.1ASGGSGSEE34 pKa = 4.07GSADD38 pKa = 3.41VDD40 pKa = 5.55EE41 pKa = 4.52ITAWAWDD48 pKa = 3.58PAFNIAALEE57 pKa = 4.13NAKK60 pKa = 9.89EE61 pKa = 4.09AYY63 pKa = 10.29NGDD66 pKa = 2.9GDD68 pKa = 4.27YY69 pKa = 11.04EE70 pKa = 4.56VNIIEE75 pKa = 4.5NAQDD79 pKa = 5.51DD80 pKa = 4.9IIQKK84 pKa = 10.54LNTGLSSGTTNGMPNIVLIEE104 pKa = 4.37DD105 pKa = 3.41YY106 pKa = 10.36RR107 pKa = 11.84AQSFLQAYY115 pKa = 8.23PDD117 pKa = 3.89AFHH120 pKa = 7.54PLTDD124 pKa = 4.61VINSDD129 pKa = 4.22DD130 pKa = 3.72FADD133 pKa = 3.78YY134 pKa = 10.8KK135 pKa = 10.78IATTSYY141 pKa = 10.91DD142 pKa = 3.31GEE144 pKa = 4.49QYY146 pKa = 11.35GLPFDD151 pKa = 4.24SGVAALYY158 pKa = 10.74VRR160 pKa = 11.84TDD162 pKa = 3.48YY163 pKa = 11.52LEE165 pKa = 4.05EE166 pKa = 5.61AGYY169 pKa = 10.44SVEE172 pKa = 4.85DD173 pKa = 3.52VTDD176 pKa = 3.66ITWQEE181 pKa = 3.99YY182 pKa = 10.34IEE184 pKa = 4.35IGKK187 pKa = 9.3DD188 pKa = 3.19VKK190 pKa = 10.77EE191 pKa = 4.02ATGKK195 pKa = 10.46NMLTLDD201 pKa = 4.5PNDD204 pKa = 3.71LAQVRR209 pKa = 11.84MMIQTNGSWYY219 pKa = 10.17VEE221 pKa = 4.27EE222 pKa = 5.93DD223 pKa = 3.24GSTPNIAGNEE233 pKa = 4.01TLEE236 pKa = 4.98AGFEE240 pKa = 4.5TYY242 pKa = 10.74KK243 pKa = 11.25EE244 pKa = 4.05MMDD247 pKa = 3.43ADD249 pKa = 3.42IAKK252 pKa = 9.81IVSDD256 pKa = 3.53WSQFVGAFNSGEE268 pKa = 4.24VASVPTGNWITPSVKK283 pKa = 9.92QAADD287 pKa = 3.49QSGKK291 pKa = 8.65WAVVPMPRR299 pKa = 11.84LDD301 pKa = 3.44MEE303 pKa = 4.69GAVNASNLGGSSWYY317 pKa = 9.91VLNVDD322 pKa = 4.65GKK324 pKa = 9.78EE325 pKa = 3.91QAADD329 pKa = 3.56FLLNTFGSNPEE340 pKa = 4.13LYY342 pKa = 8.53QTLVEE347 pKa = 5.37DD348 pKa = 3.86IGALGTYY355 pKa = 10.03SPAAEE360 pKa = 4.02GDD362 pKa = 3.47AYY364 pKa = 10.64AVEE367 pKa = 5.1DD368 pKa = 4.37EE369 pKa = 4.8FFGGQQLLTDD379 pKa = 4.45LSTWTDD385 pKa = 3.62EE386 pKa = 4.3IPAVNYY392 pKa = 10.0GLHH395 pKa = 6.22TYY397 pKa = 10.28AIEE400 pKa = 5.42DD401 pKa = 3.55ILVTGMQDD409 pKa = 3.54YY410 pKa = 11.43LNGAEE415 pKa = 4.93LDD417 pKa = 3.98TVLEE421 pKa = 4.19NVQGQAEE428 pKa = 4.28AQLKK432 pKa = 9.34
Molecular weight: 46.72 kDa
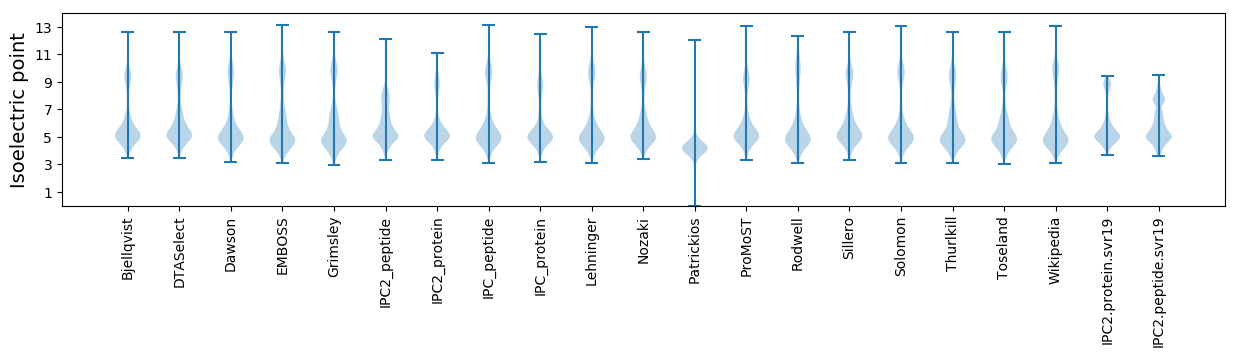
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U2Z9T2|A0A0U2Z9T2_9BACL GTPase Der OS=Planococcus rifietoensis OX=200991 GN=engA PE=3 SV=1
MM1 pKa = 7.61TLRR4 pKa = 11.84TYY6 pKa = 10.57QPNKK10 pKa = 8.74RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.91VHH17 pKa = 5.68GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.71NGRR29 pKa = 11.84RR30 pKa = 11.84VIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.8GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
MM1 pKa = 7.61TLRR4 pKa = 11.84TYY6 pKa = 10.57QPNKK10 pKa = 8.74RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.91VHH17 pKa = 5.68GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.71NGRR29 pKa = 11.84RR30 pKa = 11.84VIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.8GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
996273 |
37 |
1561 |
294.2 |
32.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.555 ± 0.041 | 0.549 ± 0.011 |
5.416 ± 0.038 | 7.962 ± 0.054 |
4.506 ± 0.037 | 7.318 ± 0.04 |
2.067 ± 0.022 | 6.948 ± 0.039 |
5.619 ± 0.035 | 9.906 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.916 ± 0.02 | 3.569 ± 0.025 |
3.82 ± 0.026 | 3.861 ± 0.028 |
4.417 ± 0.032 | 5.851 ± 0.025 |
5.337 ± 0.026 | 7.111 ± 0.028 |
1.052 ± 0.017 | 3.22 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
