
Nonomuraea maritima
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Streptosporangiaceae; Nonomuraea
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
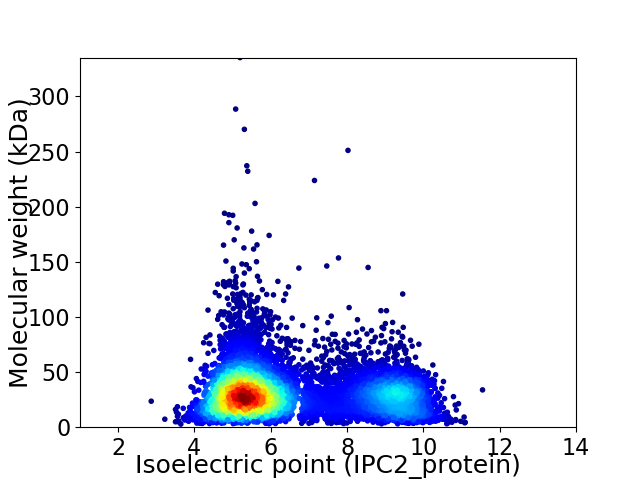
Virtual 2D-PAGE plot for 7903 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9NBA0|A0A1G9NBA0_9ACTN 1 4-dihydroxy-2-naphthoyl-CoA synthase OS=Nonomuraea maritima OX=683260 GN=menB PE=3 SV=1
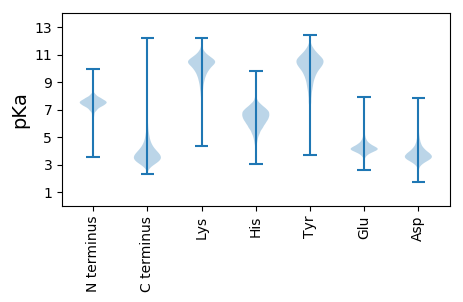
MM1 pKa = 7.8TDD3 pKa = 3.0NDD5 pKa = 4.17LTFGDD10 pKa = 4.67EE11 pKa = 4.68DD12 pKa = 4.4YY13 pKa = 11.19EE14 pKa = 4.67LPDD17 pKa = 3.25VEE19 pKa = 5.9RR20 pKa = 11.84RR21 pKa = 11.84ICPSCGTNSRR31 pKa = 11.84IITRR35 pKa = 11.84GDD37 pKa = 2.95SRR39 pKa = 11.84ATIVSKK45 pKa = 10.93CCEE48 pKa = 4.05APLSGTADD56 pKa = 3.79DD57 pKa = 5.22QDD59 pKa = 4.6DD60 pKa = 4.27EE61 pKa = 4.64PAVLVLDD68 pKa = 4.79GKK70 pKa = 9.44LVCPYY75 pKa = 10.08ADD77 pKa = 4.02CSATDD82 pKa = 4.16EE83 pKa = 4.33IVEE86 pKa = 4.32LDD88 pKa = 3.31VATRR92 pKa = 11.84SNEE95 pKa = 3.67LTIIRR100 pKa = 11.84PGVISVALGDD110 pKa = 3.88SCFEE114 pKa = 4.07NDD116 pKa = 4.06GFEE119 pKa = 4.8CEE121 pKa = 3.99QCSRR125 pKa = 11.84RR126 pKa = 11.84VSLPDD131 pKa = 3.82GYY133 pKa = 10.96EE134 pKa = 3.44LSYY137 pKa = 10.91PP138 pKa = 3.76
MM1 pKa = 7.8TDD3 pKa = 3.0NDD5 pKa = 4.17LTFGDD10 pKa = 4.67EE11 pKa = 4.68DD12 pKa = 4.4YY13 pKa = 11.19EE14 pKa = 4.67LPDD17 pKa = 3.25VEE19 pKa = 5.9RR20 pKa = 11.84RR21 pKa = 11.84ICPSCGTNSRR31 pKa = 11.84IITRR35 pKa = 11.84GDD37 pKa = 2.95SRR39 pKa = 11.84ATIVSKK45 pKa = 10.93CCEE48 pKa = 4.05APLSGTADD56 pKa = 3.79DD57 pKa = 5.22QDD59 pKa = 4.6DD60 pKa = 4.27EE61 pKa = 4.64PAVLVLDD68 pKa = 4.79GKK70 pKa = 9.44LVCPYY75 pKa = 10.08ADD77 pKa = 4.02CSATDD82 pKa = 4.16EE83 pKa = 4.33IVEE86 pKa = 4.32LDD88 pKa = 3.31VATRR92 pKa = 11.84SNEE95 pKa = 3.67LTIIRR100 pKa = 11.84PGVISVALGDD110 pKa = 3.88SCFEE114 pKa = 4.07NDD116 pKa = 4.06GFEE119 pKa = 4.8CEE121 pKa = 3.99QCSRR125 pKa = 11.84RR126 pKa = 11.84VSLPDD131 pKa = 3.82GYY133 pKa = 10.96EE134 pKa = 3.44LSYY137 pKa = 10.91PP138 pKa = 3.76
Molecular weight: 15.01 kDa
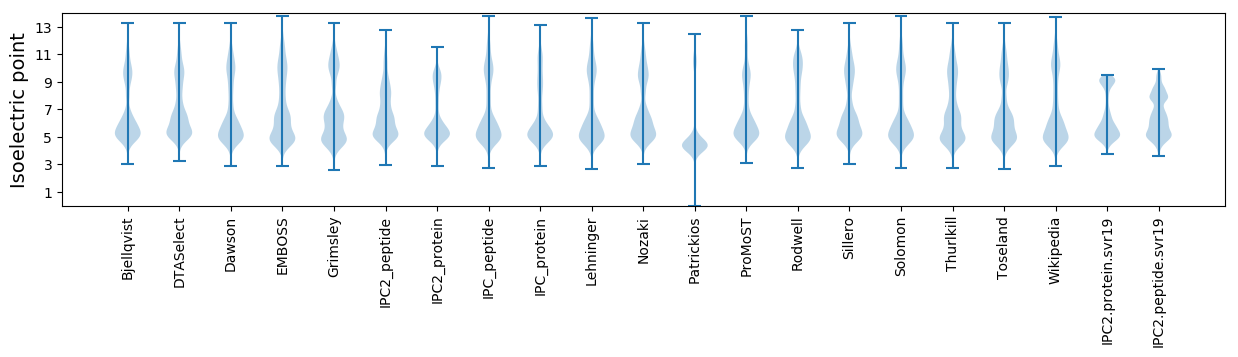
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G8UDI3|A0A1G8UDI3_9ACTN Bifunctional protein GlmU OS=Nonomuraea maritima OX=683260 GN=glmU PE=3 SV=1
MM1 pKa = 6.33QWRR4 pKa = 11.84RR5 pKa = 11.84SGYY8 pKa = 8.05TSSLRR13 pKa = 11.84SSASRR18 pKa = 11.84ARR20 pKa = 11.84SSWPSSRR27 pKa = 11.84RR28 pKa = 11.84WGSSSVRR35 pKa = 11.84RR36 pKa = 11.84PRR38 pKa = 11.84PSRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84WSAGSPKK50 pKa = 9.85LWAPRR55 pKa = 11.84RR56 pKa = 11.84VALPGAAVSRR66 pKa = 11.84PTSRR70 pKa = 11.84SRR72 pKa = 11.84QGLRR76 pKa = 11.84PGRR79 pKa = 11.84AMARR83 pKa = 11.84PATARR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84FPLRR95 pKa = 11.84VRR97 pKa = 11.84APVPVRR103 pKa = 11.84VPAPQALRR111 pKa = 11.84APQAARR117 pKa = 11.84SRR119 pKa = 11.84VRR121 pKa = 11.84RR122 pKa = 11.84FRR124 pKa = 11.84CPTSRR129 pKa = 11.84SSSPRR134 pKa = 11.84PRR136 pKa = 11.84AVRR139 pKa = 11.84PPGLRR144 pKa = 11.84RR145 pKa = 11.84RR146 pKa = 11.84ASTVARR152 pKa = 11.84PLVRR156 pKa = 11.84LRR158 pKa = 11.84ARR160 pKa = 11.84VPAPPPVPARR170 pKa = 11.84SPALGPVRR178 pKa = 11.84VLVVAVAVAVPRR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84VPARR197 pKa = 11.84LPVAPLVPARR207 pKa = 11.84APSPARR213 pKa = 11.84VVRR216 pKa = 11.84VPATTRR222 pKa = 11.84SRR224 pKa = 11.84PTRR227 pKa = 11.84AAWVSRR233 pKa = 11.84VRR235 pKa = 11.84RR236 pKa = 11.84VPVATAKK243 pKa = 10.54AAAPASASVVTVRR256 pKa = 11.84RR257 pKa = 11.84ATARR261 pKa = 11.84CRR263 pKa = 11.84VRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84AVPAPTPGPAPAARR281 pKa = 11.84VPARR285 pKa = 11.84RR286 pKa = 11.84PAVRR290 pKa = 11.84VRR292 pKa = 11.84VPVRR296 pKa = 11.84VVRR299 pKa = 11.84VRR301 pKa = 11.84VARR304 pKa = 11.84VPTRR308 pKa = 3.2
MM1 pKa = 6.33QWRR4 pKa = 11.84RR5 pKa = 11.84SGYY8 pKa = 8.05TSSLRR13 pKa = 11.84SSASRR18 pKa = 11.84ARR20 pKa = 11.84SSWPSSRR27 pKa = 11.84RR28 pKa = 11.84WGSSSVRR35 pKa = 11.84RR36 pKa = 11.84PRR38 pKa = 11.84PSRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84WSAGSPKK50 pKa = 9.85LWAPRR55 pKa = 11.84RR56 pKa = 11.84VALPGAAVSRR66 pKa = 11.84PTSRR70 pKa = 11.84SRR72 pKa = 11.84QGLRR76 pKa = 11.84PGRR79 pKa = 11.84AMARR83 pKa = 11.84PATARR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84FPLRR95 pKa = 11.84VRR97 pKa = 11.84APVPVRR103 pKa = 11.84VPAPQALRR111 pKa = 11.84APQAARR117 pKa = 11.84SRR119 pKa = 11.84VRR121 pKa = 11.84RR122 pKa = 11.84FRR124 pKa = 11.84CPTSRR129 pKa = 11.84SSSPRR134 pKa = 11.84PRR136 pKa = 11.84AVRR139 pKa = 11.84PPGLRR144 pKa = 11.84RR145 pKa = 11.84RR146 pKa = 11.84ASTVARR152 pKa = 11.84PLVRR156 pKa = 11.84LRR158 pKa = 11.84ARR160 pKa = 11.84VPAPPPVPARR170 pKa = 11.84SPALGPVRR178 pKa = 11.84VLVVAVAVAVPRR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84VPARR197 pKa = 11.84LPVAPLVPARR207 pKa = 11.84APSPARR213 pKa = 11.84VVRR216 pKa = 11.84VPATTRR222 pKa = 11.84SRR224 pKa = 11.84PTRR227 pKa = 11.84AAWVSRR233 pKa = 11.84VRR235 pKa = 11.84RR236 pKa = 11.84VPVATAKK243 pKa = 10.54AAAPASASVVTVRR256 pKa = 11.84RR257 pKa = 11.84ATARR261 pKa = 11.84CRR263 pKa = 11.84VRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84AVPAPTPGPAPAARR281 pKa = 11.84VPARR285 pKa = 11.84RR286 pKa = 11.84PAVRR290 pKa = 11.84VRR292 pKa = 11.84VPVRR296 pKa = 11.84VVRR299 pKa = 11.84VRR301 pKa = 11.84VARR304 pKa = 11.84VPTRR308 pKa = 3.2
Molecular weight: 33.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2450462 |
26 |
3110 |
310.1 |
33.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.225 ± 0.04 | 0.78 ± 0.009 |
5.887 ± 0.022 | 5.683 ± 0.025 |
2.8 ± 0.015 | 9.189 ± 0.028 |
2.272 ± 0.011 | 3.426 ± 0.018 |
2.018 ± 0.019 | 10.591 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.935 ± 0.012 | 1.722 ± 0.013 |
6.042 ± 0.026 | 2.7 ± 0.016 |
8.302 ± 0.03 | 4.954 ± 0.02 |
5.806 ± 0.021 | 9.015 ± 0.027 |
1.548 ± 0.011 | 2.106 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
