
Pacific flying fox faeces associated circular DNA virus-6
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.19
Get precalculated fractions of proteins
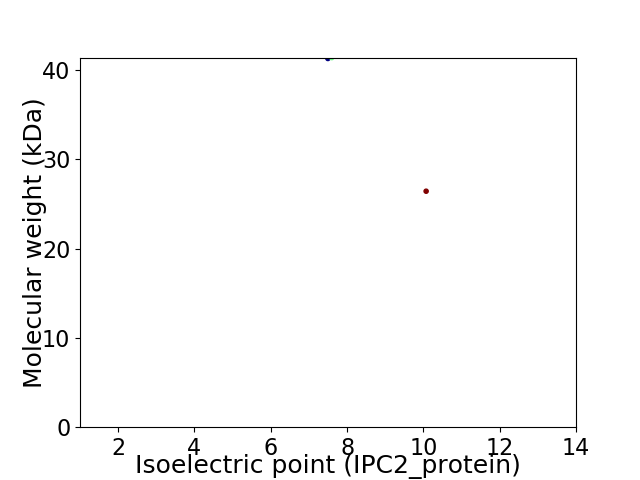
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A140CTU1|A0A140CTU1_9VIRU ATP-dependent helicase Rep OS=Pacific flying fox faeces associated circular DNA virus-6 OX=1796015 PE=3 SV=1
MM1 pKa = 7.46SRR3 pKa = 11.84DD4 pKa = 3.77KK5 pKa = 11.47VDD7 pKa = 4.01LAPSRR12 pKa = 11.84GFSKK16 pKa = 10.32YY17 pKa = 6.88WCFTAFASDD26 pKa = 3.75FEE28 pKa = 4.67KK29 pKa = 10.3FTEE32 pKa = 4.77IISDD36 pKa = 3.29EE37 pKa = 4.29RR38 pKa = 11.84VFSNSDD44 pKa = 2.85LSYY47 pKa = 10.57IIGGEE52 pKa = 4.1EE53 pKa = 4.13LCPLSGRR60 pKa = 11.84KK61 pKa = 9.13HH62 pKa = 5.0IQGFAAFASRR72 pKa = 11.84KK73 pKa = 9.66RR74 pKa = 11.84LTACKK79 pKa = 10.12KK80 pKa = 9.74LFPSAHH86 pKa = 6.09FEE88 pKa = 4.05RR89 pKa = 11.84MRR91 pKa = 11.84GSARR95 pKa = 11.84EE96 pKa = 3.95AIEE99 pKa = 3.88YY100 pKa = 9.34CKK102 pKa = 10.52KK103 pKa = 10.47DD104 pKa = 3.17GKK106 pKa = 10.5FVEE109 pKa = 4.42RR110 pKa = 11.84GIYY113 pKa = 9.05PCVRR117 pKa = 11.84SSTVDD122 pKa = 2.9NGSAKK127 pKa = 10.47GVDD130 pKa = 3.54RR131 pKa = 11.84IKK133 pKa = 11.24DD134 pKa = 4.08FIEE137 pKa = 3.8LARR140 pKa = 11.84AGNVDD145 pKa = 3.81LCEE148 pKa = 3.96QRR150 pKa = 11.84HH151 pKa = 5.55PNQFLRR157 pKa = 11.84YY158 pKa = 8.89FSQLKK163 pKa = 9.76NLRR166 pKa = 11.84KK167 pKa = 9.42FDD169 pKa = 3.64IEE171 pKa = 4.18RR172 pKa = 11.84LSEE175 pKa = 4.06ALWRR179 pKa = 11.84LDD181 pKa = 3.38LRR183 pKa = 11.84GPSFGIGKK191 pKa = 9.35DD192 pKa = 3.41ASRR195 pKa = 11.84SPSLRR200 pKa = 11.84VFFFKK205 pKa = 10.61KK206 pKa = 10.32SQLKK210 pKa = 8.5WWDD213 pKa = 3.49GYY215 pKa = 11.3NNEE218 pKa = 4.35NIILISDD225 pKa = 4.15FDD227 pKa = 4.66EE228 pKa = 4.35SCKK231 pKa = 10.62WMVHH235 pKa = 5.09YY236 pKa = 10.41LKK238 pKa = 10.06IWADD242 pKa = 3.3VYY244 pKa = 11.05PFNAEE249 pKa = 4.21VKK251 pKa = 10.39GSTIKK256 pKa = 10.19IRR258 pKa = 11.84PKK260 pKa = 10.61YY261 pKa = 10.38IIVTSNFRR269 pKa = 11.84MDD271 pKa = 3.52MLFLGTALDD280 pKa = 3.62ALKK283 pKa = 10.52RR284 pKa = 11.84RR285 pKa = 11.84FMTFEE290 pKa = 3.83FSGSGLVEE298 pKa = 3.62KK299 pKa = 10.04QLRR302 pKa = 11.84NEE304 pKa = 3.92SHH306 pKa = 6.96FNAFVLDD313 pKa = 3.77ALKK316 pKa = 10.41PYY318 pKa = 10.49VDD320 pKa = 4.44EE321 pKa = 5.98DD322 pKa = 3.73VDD324 pKa = 4.06ASPSPSSSVLPNGGEE339 pKa = 3.61ISPFTDD345 pKa = 3.7EE346 pKa = 4.43EE347 pKa = 4.86TSTCLPSTSCAVDD360 pKa = 3.32NPP362 pKa = 3.95
MM1 pKa = 7.46SRR3 pKa = 11.84DD4 pKa = 3.77KK5 pKa = 11.47VDD7 pKa = 4.01LAPSRR12 pKa = 11.84GFSKK16 pKa = 10.32YY17 pKa = 6.88WCFTAFASDD26 pKa = 3.75FEE28 pKa = 4.67KK29 pKa = 10.3FTEE32 pKa = 4.77IISDD36 pKa = 3.29EE37 pKa = 4.29RR38 pKa = 11.84VFSNSDD44 pKa = 2.85LSYY47 pKa = 10.57IIGGEE52 pKa = 4.1EE53 pKa = 4.13LCPLSGRR60 pKa = 11.84KK61 pKa = 9.13HH62 pKa = 5.0IQGFAAFASRR72 pKa = 11.84KK73 pKa = 9.66RR74 pKa = 11.84LTACKK79 pKa = 10.12KK80 pKa = 9.74LFPSAHH86 pKa = 6.09FEE88 pKa = 4.05RR89 pKa = 11.84MRR91 pKa = 11.84GSARR95 pKa = 11.84EE96 pKa = 3.95AIEE99 pKa = 3.88YY100 pKa = 9.34CKK102 pKa = 10.52KK103 pKa = 10.47DD104 pKa = 3.17GKK106 pKa = 10.5FVEE109 pKa = 4.42RR110 pKa = 11.84GIYY113 pKa = 9.05PCVRR117 pKa = 11.84SSTVDD122 pKa = 2.9NGSAKK127 pKa = 10.47GVDD130 pKa = 3.54RR131 pKa = 11.84IKK133 pKa = 11.24DD134 pKa = 4.08FIEE137 pKa = 3.8LARR140 pKa = 11.84AGNVDD145 pKa = 3.81LCEE148 pKa = 3.96QRR150 pKa = 11.84HH151 pKa = 5.55PNQFLRR157 pKa = 11.84YY158 pKa = 8.89FSQLKK163 pKa = 9.76NLRR166 pKa = 11.84KK167 pKa = 9.42FDD169 pKa = 3.64IEE171 pKa = 4.18RR172 pKa = 11.84LSEE175 pKa = 4.06ALWRR179 pKa = 11.84LDD181 pKa = 3.38LRR183 pKa = 11.84GPSFGIGKK191 pKa = 9.35DD192 pKa = 3.41ASRR195 pKa = 11.84SPSLRR200 pKa = 11.84VFFFKK205 pKa = 10.61KK206 pKa = 10.32SQLKK210 pKa = 8.5WWDD213 pKa = 3.49GYY215 pKa = 11.3NNEE218 pKa = 4.35NIILISDD225 pKa = 4.15FDD227 pKa = 4.66EE228 pKa = 4.35SCKK231 pKa = 10.62WMVHH235 pKa = 5.09YY236 pKa = 10.41LKK238 pKa = 10.06IWADD242 pKa = 3.3VYY244 pKa = 11.05PFNAEE249 pKa = 4.21VKK251 pKa = 10.39GSTIKK256 pKa = 10.19IRR258 pKa = 11.84PKK260 pKa = 10.61YY261 pKa = 10.38IIVTSNFRR269 pKa = 11.84MDD271 pKa = 3.52MLFLGTALDD280 pKa = 3.62ALKK283 pKa = 10.52RR284 pKa = 11.84RR285 pKa = 11.84FMTFEE290 pKa = 3.83FSGSGLVEE298 pKa = 3.62KK299 pKa = 10.04QLRR302 pKa = 11.84NEE304 pKa = 3.92SHH306 pKa = 6.96FNAFVLDD313 pKa = 3.77ALKK316 pKa = 10.41PYY318 pKa = 10.49VDD320 pKa = 4.44EE321 pKa = 5.98DD322 pKa = 3.73VDD324 pKa = 4.06ASPSPSSSVLPNGGEE339 pKa = 3.61ISPFTDD345 pKa = 3.7EE346 pKa = 4.43EE347 pKa = 4.86TSTCLPSTSCAVDD360 pKa = 3.32NPP362 pKa = 3.95
Molecular weight: 41.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTU1|A0A140CTU1_9VIRU ATP-dependent helicase Rep OS=Pacific flying fox faeces associated circular DNA virus-6 OX=1796015 PE=3 SV=1
MM1 pKa = 7.6WMRR4 pKa = 11.84MWMRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PFYY17 pKa = 9.89RR18 pKa = 11.84TAAKK22 pKa = 10.08YY23 pKa = 10.04RR24 pKa = 11.84RR25 pKa = 11.84LRR27 pKa = 11.84MKK29 pKa = 9.79KK30 pKa = 9.2RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84VYY35 pKa = 9.47HH36 pKa = 6.28RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84VPLIIRR45 pKa = 11.84EE46 pKa = 4.18NYY48 pKa = 9.47KK49 pKa = 9.02KK50 pKa = 9.42TIVRR54 pKa = 11.84KK55 pKa = 8.21VDD57 pKa = 2.83QWNTHH62 pKa = 3.81VWRR65 pKa = 11.84GIRR68 pKa = 11.84SVAPDD73 pKa = 2.85NFMRR77 pKa = 11.84FRR79 pKa = 11.84DD80 pKa = 3.7KK81 pKa = 10.94LRR83 pKa = 11.84CEE85 pKa = 4.66MLTQDD90 pKa = 4.03PQFISDD96 pKa = 3.76LADD99 pKa = 3.16YY100 pKa = 10.72KK101 pKa = 10.9YY102 pKa = 11.16FKK104 pKa = 11.18LNWFKK109 pKa = 11.38LKK111 pKa = 10.14IVSIAALEE119 pKa = 4.02QQAMRR124 pKa = 11.84VPPNSDD130 pKa = 2.84PTLGFKK136 pKa = 10.49SVIGHH141 pKa = 6.55TGMNSLAWYY150 pKa = 9.84VDD152 pKa = 2.71WDD154 pKa = 4.28LDD156 pKa = 4.12HH157 pKa = 7.36IAPKK161 pKa = 10.52SPEE164 pKa = 4.03DD165 pKa = 3.22LRR167 pKa = 11.84QRR169 pKa = 11.84VSSYY173 pKa = 10.9NMKK176 pKa = 10.41KK177 pKa = 9.62MYY179 pKa = 10.24VGSFKK184 pKa = 10.68PVTLFYY190 pKa = 10.81RR191 pKa = 11.84LPAEE195 pKa = 3.93CRR197 pKa = 11.84MYY199 pKa = 10.81RR200 pKa = 11.84ATALTASSGWKK211 pKa = 9.57PSEE214 pKa = 4.07STIGG218 pKa = 3.29
MM1 pKa = 7.6WMRR4 pKa = 11.84MWMRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PFYY17 pKa = 9.89RR18 pKa = 11.84TAAKK22 pKa = 10.08YY23 pKa = 10.04RR24 pKa = 11.84RR25 pKa = 11.84LRR27 pKa = 11.84MKK29 pKa = 9.79KK30 pKa = 9.2RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84VYY35 pKa = 9.47HH36 pKa = 6.28RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84VPLIIRR45 pKa = 11.84EE46 pKa = 4.18NYY48 pKa = 9.47KK49 pKa = 9.02KK50 pKa = 9.42TIVRR54 pKa = 11.84KK55 pKa = 8.21VDD57 pKa = 2.83QWNTHH62 pKa = 3.81VWRR65 pKa = 11.84GIRR68 pKa = 11.84SVAPDD73 pKa = 2.85NFMRR77 pKa = 11.84FRR79 pKa = 11.84DD80 pKa = 3.7KK81 pKa = 10.94LRR83 pKa = 11.84CEE85 pKa = 4.66MLTQDD90 pKa = 4.03PQFISDD96 pKa = 3.76LADD99 pKa = 3.16YY100 pKa = 10.72KK101 pKa = 10.9YY102 pKa = 11.16FKK104 pKa = 11.18LNWFKK109 pKa = 11.38LKK111 pKa = 10.14IVSIAALEE119 pKa = 4.02QQAMRR124 pKa = 11.84VPPNSDD130 pKa = 2.84PTLGFKK136 pKa = 10.49SVIGHH141 pKa = 6.55TGMNSLAWYY150 pKa = 9.84VDD152 pKa = 2.71WDD154 pKa = 4.28LDD156 pKa = 4.12HH157 pKa = 7.36IAPKK161 pKa = 10.52SPEE164 pKa = 4.03DD165 pKa = 3.22LRR167 pKa = 11.84QRR169 pKa = 11.84VSSYY173 pKa = 10.9NMKK176 pKa = 10.41KK177 pKa = 9.62MYY179 pKa = 10.24VGSFKK184 pKa = 10.68PVTLFYY190 pKa = 10.81RR191 pKa = 11.84LPAEE195 pKa = 3.93CRR197 pKa = 11.84MYY199 pKa = 10.81RR200 pKa = 11.84ATALTASSGWKK211 pKa = 9.57PSEE214 pKa = 4.07STIGG218 pKa = 3.29
Molecular weight: 26.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
580 |
218 |
362 |
290.0 |
33.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.207 ± 0.14 | 1.897 ± 0.563 |
6.207 ± 0.668 | 5.0 ± 1.293 |
6.552 ± 1.394 | 4.828 ± 0.93 |
1.552 ± 0.163 | 5.172 ± 0.337 |
7.414 ± 0.221 | 7.586 ± 0.406 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.103 ± 1.381 | 3.621 ± 0.236 |
4.828 ± 0.389 | 2.069 ± 0.393 |
9.828 ± 2.527 | 8.966 ± 1.463 |
3.793 ± 0.457 | 5.345 ± 0.356 |
2.414 ± 0.722 | 3.621 ± 0.82 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
