
Gorilla anellovirus
Taxonomy: Viruses; Anelloviridae; Omegatorquevirus; Torque teno hominid virus 1
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
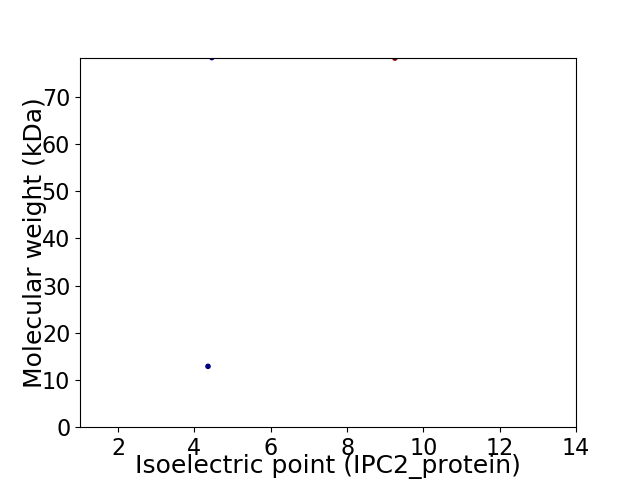
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2GN10|A0A0S2GN10_9VIRU ORF2 OS=Gorilla anellovirus OX=1743411 PE=4 SV=1
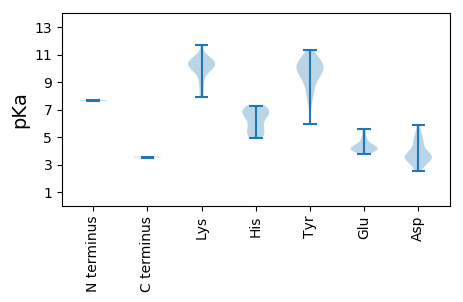
MM1 pKa = 7.63SDD3 pKa = 2.66RR4 pKa = 11.84WIPPKK9 pKa = 10.45YY10 pKa = 8.96AWQGRR15 pKa = 11.84EE16 pKa = 4.06LQWINSIHH24 pKa = 5.12TTHH27 pKa = 6.9DD28 pKa = 3.3TWCGCSSVITHH39 pKa = 6.95FLRR42 pKa = 11.84AVSARR47 pKa = 11.84GEE49 pKa = 4.11FLPVYY54 pKa = 9.51TPAEE58 pKa = 4.06KK59 pKa = 10.32AISFLPEE66 pKa = 4.15CPTTTEE72 pKa = 4.43EE73 pKa = 4.4IPPITATGDD82 pKa = 3.05TSGIHH87 pKa = 5.09TEE89 pKa = 4.14EE90 pKa = 3.94EE91 pKa = 3.97RR92 pKa = 11.84GLLDD96 pKa = 5.02LEE98 pKa = 5.05EE99 pKa = 5.57GDD101 pKa = 4.67SDD103 pKa = 5.23ALFADD108 pKa = 4.04DD109 pKa = 5.28HH110 pKa = 7.07EE111 pKa = 4.82EE112 pKa = 4.15DD113 pKa = 3.69AAGG116 pKa = 3.58
MM1 pKa = 7.63SDD3 pKa = 2.66RR4 pKa = 11.84WIPPKK9 pKa = 10.45YY10 pKa = 8.96AWQGRR15 pKa = 11.84EE16 pKa = 4.06LQWINSIHH24 pKa = 5.12TTHH27 pKa = 6.9DD28 pKa = 3.3TWCGCSSVITHH39 pKa = 6.95FLRR42 pKa = 11.84AVSARR47 pKa = 11.84GEE49 pKa = 4.11FLPVYY54 pKa = 9.51TPAEE58 pKa = 4.06KK59 pKa = 10.32AISFLPEE66 pKa = 4.15CPTTTEE72 pKa = 4.43EE73 pKa = 4.4IPPITATGDD82 pKa = 3.05TSGIHH87 pKa = 5.09TEE89 pKa = 4.14EE90 pKa = 3.94EE91 pKa = 3.97RR92 pKa = 11.84GLLDD96 pKa = 5.02LEE98 pKa = 5.05EE99 pKa = 5.57GDD101 pKa = 4.67SDD103 pKa = 5.23ALFADD108 pKa = 4.04DD109 pKa = 5.28HH110 pKa = 7.07EE111 pKa = 4.82EE112 pKa = 4.15DD113 pKa = 3.69AAGG116 pKa = 3.58
Molecular weight: 12.89 kDa
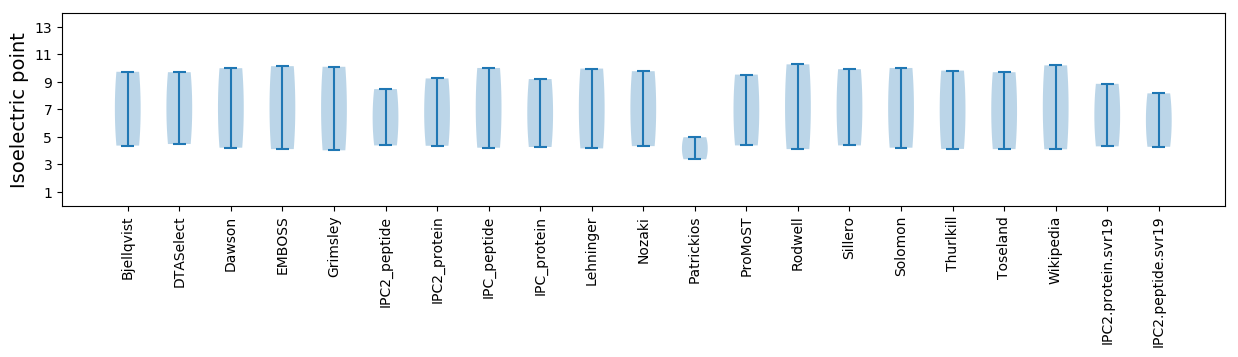
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2GN10|A0A0S2GN10_9VIRU ORF2 OS=Gorilla anellovirus OX=1743411 PE=4 SV=1
MM1 pKa = 7.71PYY3 pKa = 10.06YY4 pKa = 10.5YY5 pKa = 10.23RR6 pKa = 11.84RR7 pKa = 11.84NTPYY11 pKa = 10.05YY12 pKa = 9.62RR13 pKa = 11.84YY14 pKa = 9.82RR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 9.32FWNPYY22 pKa = 8.16RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 10.04RR27 pKa = 11.84IIRR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84RR38 pKa = 11.84LVRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84PRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84WVRR52 pKa = 11.84RR53 pKa = 11.84KK54 pKa = 10.28KK55 pKa = 9.62KK56 pKa = 9.26TIFLKK61 pKa = 9.91QWQPPTIRR69 pKa = 11.84RR70 pKa = 11.84CKK72 pKa = 10.12IKK74 pKa = 10.82GHH76 pKa = 5.71MCSMMCAHH84 pKa = 6.9GNEE87 pKa = 4.12SHH89 pKa = 7.28NYY91 pKa = 7.24NQYY94 pKa = 9.61VKK96 pKa = 10.51SYY98 pKa = 9.72TPSNYY103 pKa = 9.49PGGGGFSILRR113 pKa = 11.84FSLGALYY120 pKa = 10.22EE121 pKa = 4.13QFSDD125 pKa = 5.85FFNFWTKK132 pKa = 7.89TNKK135 pKa = 9.94NLDD138 pKa = 3.22LCRR141 pKa = 11.84YY142 pKa = 8.49IKK144 pKa = 10.83CNLLFWRR151 pKa = 11.84HH152 pKa = 5.45SDD154 pKa = 2.89VSYY157 pKa = 10.54IVSYY161 pKa = 10.39NKK163 pKa = 10.08NYY165 pKa = 10.12PMTANPLSHH174 pKa = 6.59AQAHH178 pKa = 6.59PSRR181 pKa = 11.84QILLKK186 pKa = 10.39KK187 pKa = 10.27KK188 pKa = 10.58LIVPSFKK195 pKa = 10.57EE196 pKa = 3.79NRR198 pKa = 11.84NKK200 pKa = 10.38KK201 pKa = 8.37PYY203 pKa = 9.94KK204 pKa = 10.0RR205 pKa = 11.84LTIKK209 pKa = 10.5PPSLLNSKK217 pKa = 8.67WFFQKK222 pKa = 10.75DD223 pKa = 3.44FADD226 pKa = 3.34VGLFLLTVTSCSLNHH241 pKa = 6.68AFAATNAEE249 pKa = 4.3SNNISIYY256 pKa = 10.6CLNTQNVFKK265 pKa = 10.54KK266 pKa = 9.94INYY269 pKa = 5.92VTTSGQQNYY278 pKa = 8.44QPLGGTEE285 pKa = 4.18TLFAASQKK293 pKa = 9.87TNLTDD298 pKa = 3.39NNTWPLGSQKK308 pKa = 9.0YY309 pKa = 7.84TEE311 pKa = 5.0GNPPSNPGNIFFKK324 pKa = 10.54SYY326 pKa = 11.33ASGEE330 pKa = 4.22TYY332 pKa = 10.06IFKK335 pKa = 10.74GNKK338 pKa = 8.78PKK340 pKa = 9.91GTEE343 pKa = 4.05FKK345 pKa = 10.15EE346 pKa = 3.78PLYY349 pKa = 10.43IEE351 pKa = 4.93CRR353 pKa = 11.84YY354 pKa = 10.66NPLLDD359 pKa = 4.61NGDD362 pKa = 3.59GNQIYY367 pKa = 10.41LLSTGTTSNPWQPPSDD383 pKa = 3.54QQLIISGLPLYY394 pKa = 10.06IALFGWTDD402 pKa = 3.88WIRR405 pKa = 11.84KK406 pKa = 7.91LRR408 pKa = 11.84PASQIDD414 pKa = 3.46INYY417 pKa = 8.87QLVIITDD424 pKa = 4.56KK425 pKa = 10.27ITPKK429 pKa = 10.42LPAYY433 pKa = 9.66IPISEE438 pKa = 4.27YY439 pKa = 10.58FIEE442 pKa = 5.39GNGLYY447 pKa = 9.04QQPLTDD453 pKa = 3.5TEE455 pKa = 4.19LAHH458 pKa = 6.91WYY460 pKa = 9.44PRR462 pKa = 11.84VKK464 pKa = 10.48FQEE467 pKa = 3.83PAINDD472 pKa = 3.41IVKK475 pKa = 10.15CGPFMPRR482 pKa = 11.84AEE484 pKa = 5.32GIHH487 pKa = 5.97SWEE490 pKa = 3.91AHH492 pKa = 4.91CTYY495 pKa = 11.24NFIFKK500 pKa = 9.56WGGSVPPEE508 pKa = 3.9QDD510 pKa = 2.51IKK512 pKa = 11.68DD513 pKa = 4.76PISQPAYY520 pKa = 10.12SVPDD524 pKa = 3.72KK525 pKa = 11.16LSTTIQIEE533 pKa = 4.24DD534 pKa = 3.71PRR536 pKa = 11.84NISPYY541 pKa = 7.95TTLHH545 pKa = 5.88PWDD548 pKa = 3.63IRR550 pKa = 11.84RR551 pKa = 11.84GTLTEE556 pKa = 4.1TTLKK560 pKa = 10.6RR561 pKa = 11.84ILEE564 pKa = 4.35DD565 pKa = 3.72AEE567 pKa = 4.65TKK569 pKa = 10.54ASCITGNEE577 pKa = 4.02STQILTKK584 pKa = 10.13RR585 pKa = 11.84RR586 pKa = 11.84RR587 pKa = 11.84TMEE590 pKa = 4.74PDD592 pKa = 3.78LYY594 pKa = 10.99TDD596 pKa = 4.35IPWDD600 pKa = 3.44QQLLHH605 pKa = 6.81KK606 pKa = 10.67AFEE609 pKa = 4.28EE610 pKa = 4.47NIFQEE615 pKa = 4.49TEE617 pKa = 4.03EE618 pKa = 4.49EE619 pKa = 4.34EE620 pKa = 4.44TTQGNLIEE628 pKa = 4.4QFNKK632 pKa = 9.86QKK634 pKa = 10.04QQQQRR639 pKa = 11.84LRR641 pKa = 11.84QQLTTVILSLKK652 pKa = 8.09EE653 pKa = 4.05QQHH656 pKa = 5.22KK657 pKa = 9.16MQMQMGTLL665 pKa = 3.51
MM1 pKa = 7.71PYY3 pKa = 10.06YY4 pKa = 10.5YY5 pKa = 10.23RR6 pKa = 11.84RR7 pKa = 11.84NTPYY11 pKa = 10.05YY12 pKa = 9.62RR13 pKa = 11.84YY14 pKa = 9.82RR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 9.32FWNPYY22 pKa = 8.16RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 10.04RR27 pKa = 11.84IIRR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84RR38 pKa = 11.84LVRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84PRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84WVRR52 pKa = 11.84RR53 pKa = 11.84KK54 pKa = 10.28KK55 pKa = 9.62KK56 pKa = 9.26TIFLKK61 pKa = 9.91QWQPPTIRR69 pKa = 11.84RR70 pKa = 11.84CKK72 pKa = 10.12IKK74 pKa = 10.82GHH76 pKa = 5.71MCSMMCAHH84 pKa = 6.9GNEE87 pKa = 4.12SHH89 pKa = 7.28NYY91 pKa = 7.24NQYY94 pKa = 9.61VKK96 pKa = 10.51SYY98 pKa = 9.72TPSNYY103 pKa = 9.49PGGGGFSILRR113 pKa = 11.84FSLGALYY120 pKa = 10.22EE121 pKa = 4.13QFSDD125 pKa = 5.85FFNFWTKK132 pKa = 7.89TNKK135 pKa = 9.94NLDD138 pKa = 3.22LCRR141 pKa = 11.84YY142 pKa = 8.49IKK144 pKa = 10.83CNLLFWRR151 pKa = 11.84HH152 pKa = 5.45SDD154 pKa = 2.89VSYY157 pKa = 10.54IVSYY161 pKa = 10.39NKK163 pKa = 10.08NYY165 pKa = 10.12PMTANPLSHH174 pKa = 6.59AQAHH178 pKa = 6.59PSRR181 pKa = 11.84QILLKK186 pKa = 10.39KK187 pKa = 10.27KK188 pKa = 10.58LIVPSFKK195 pKa = 10.57EE196 pKa = 3.79NRR198 pKa = 11.84NKK200 pKa = 10.38KK201 pKa = 8.37PYY203 pKa = 9.94KK204 pKa = 10.0RR205 pKa = 11.84LTIKK209 pKa = 10.5PPSLLNSKK217 pKa = 8.67WFFQKK222 pKa = 10.75DD223 pKa = 3.44FADD226 pKa = 3.34VGLFLLTVTSCSLNHH241 pKa = 6.68AFAATNAEE249 pKa = 4.3SNNISIYY256 pKa = 10.6CLNTQNVFKK265 pKa = 10.54KK266 pKa = 9.94INYY269 pKa = 5.92VTTSGQQNYY278 pKa = 8.44QPLGGTEE285 pKa = 4.18TLFAASQKK293 pKa = 9.87TNLTDD298 pKa = 3.39NNTWPLGSQKK308 pKa = 9.0YY309 pKa = 7.84TEE311 pKa = 5.0GNPPSNPGNIFFKK324 pKa = 10.54SYY326 pKa = 11.33ASGEE330 pKa = 4.22TYY332 pKa = 10.06IFKK335 pKa = 10.74GNKK338 pKa = 8.78PKK340 pKa = 9.91GTEE343 pKa = 4.05FKK345 pKa = 10.15EE346 pKa = 3.78PLYY349 pKa = 10.43IEE351 pKa = 4.93CRR353 pKa = 11.84YY354 pKa = 10.66NPLLDD359 pKa = 4.61NGDD362 pKa = 3.59GNQIYY367 pKa = 10.41LLSTGTTSNPWQPPSDD383 pKa = 3.54QQLIISGLPLYY394 pKa = 10.06IALFGWTDD402 pKa = 3.88WIRR405 pKa = 11.84KK406 pKa = 7.91LRR408 pKa = 11.84PASQIDD414 pKa = 3.46INYY417 pKa = 8.87QLVIITDD424 pKa = 4.56KK425 pKa = 10.27ITPKK429 pKa = 10.42LPAYY433 pKa = 9.66IPISEE438 pKa = 4.27YY439 pKa = 10.58FIEE442 pKa = 5.39GNGLYY447 pKa = 9.04QQPLTDD453 pKa = 3.5TEE455 pKa = 4.19LAHH458 pKa = 6.91WYY460 pKa = 9.44PRR462 pKa = 11.84VKK464 pKa = 10.48FQEE467 pKa = 3.83PAINDD472 pKa = 3.41IVKK475 pKa = 10.15CGPFMPRR482 pKa = 11.84AEE484 pKa = 5.32GIHH487 pKa = 5.97SWEE490 pKa = 3.91AHH492 pKa = 4.91CTYY495 pKa = 11.24NFIFKK500 pKa = 9.56WGGSVPPEE508 pKa = 3.9QDD510 pKa = 2.51IKK512 pKa = 11.68DD513 pKa = 4.76PISQPAYY520 pKa = 10.12SVPDD524 pKa = 3.72KK525 pKa = 11.16LSTTIQIEE533 pKa = 4.24DD534 pKa = 3.71PRR536 pKa = 11.84NISPYY541 pKa = 7.95TTLHH545 pKa = 5.88PWDD548 pKa = 3.63IRR550 pKa = 11.84RR551 pKa = 11.84GTLTEE556 pKa = 4.1TTLKK560 pKa = 10.6RR561 pKa = 11.84ILEE564 pKa = 4.35DD565 pKa = 3.72AEE567 pKa = 4.65TKK569 pKa = 10.54ASCITGNEE577 pKa = 4.02STQILTKK584 pKa = 10.13RR585 pKa = 11.84RR586 pKa = 11.84RR587 pKa = 11.84TMEE590 pKa = 4.74PDD592 pKa = 3.78LYY594 pKa = 10.99TDD596 pKa = 4.35IPWDD600 pKa = 3.44QQLLHH605 pKa = 6.81KK606 pKa = 10.67AFEE609 pKa = 4.28EE610 pKa = 4.47NIFQEE615 pKa = 4.49TEE617 pKa = 4.03EE618 pKa = 4.49EE619 pKa = 4.34EE620 pKa = 4.44TTQGNLIEE628 pKa = 4.4QFNKK632 pKa = 9.86QKK634 pKa = 10.04QQQQRR639 pKa = 11.84LRR641 pKa = 11.84QQLTTVILSLKK652 pKa = 8.09EE653 pKa = 4.05QQHH656 pKa = 5.22KK657 pKa = 9.16MQMQMGTLL665 pKa = 3.51
Molecular weight: 78.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
781 |
116 |
665 |
390.5 |
45.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.353 ± 1.988 | 1.793 ± 0.37 |
4.097 ± 1.706 | 5.762 ± 2.537 |
4.353 ± 0.422 | 5.122 ± 0.827 |
2.305 ± 0.934 | 7.042 ± 0.068 |
6.274 ± 2.12 | 8.195 ± 0.605 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.408 ± 0.255 | 5.762 ± 2.283 |
7.042 ± 0.068 | 5.634 ± 1.821 |
6.786 ± 1.153 | 6.146 ± 0.35 |
8.067 ± 1.061 | 2.433 ± 0.071 |
2.433 ± 0.473 | 4.994 ± 1.523 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
