
Sanxia water strider virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
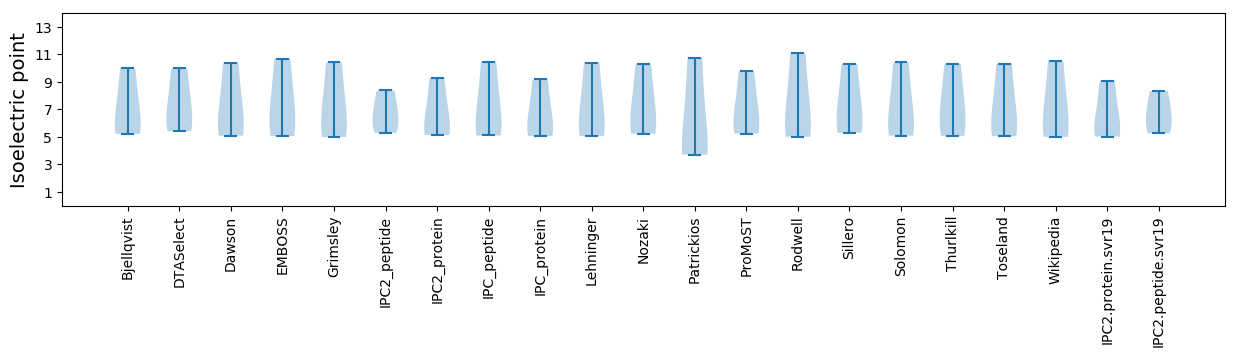
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
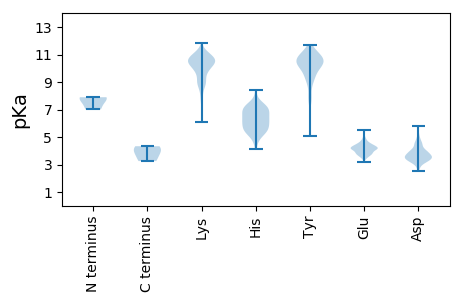
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJ76|A0A1L3KJ76_9VIRU Uncharacterized protein OS=Sanxia water strider virus 7 OX=1923406 PE=4 SV=1
MM1 pKa = 7.82SDD3 pKa = 3.27PHH5 pKa = 7.31GDD7 pKa = 3.49PAPVDD12 pKa = 3.38RR13 pKa = 11.84GGVISVEE20 pKa = 4.13VSEE23 pKa = 4.61EE24 pKa = 4.21SSGKK28 pKa = 9.5PDD30 pKa = 3.71FPTSIILSRR39 pKa = 11.84FCSVEE44 pKa = 3.77SEE46 pKa = 4.23QVYY49 pKa = 10.9KK50 pKa = 11.07DD51 pKa = 4.5LLNTWSILPPIIWEE65 pKa = 4.3GTTDD69 pKa = 4.98AVTTYY74 pKa = 10.53NLPYY78 pKa = 10.26YY79 pKa = 10.4AVEE82 pKa = 4.13SLKK85 pKa = 10.55DD86 pKa = 3.89YY87 pKa = 10.94PQMLPFRR94 pKa = 11.84MYY96 pKa = 10.23QYY98 pKa = 9.9WRR100 pKa = 11.84GDD102 pKa = 3.12IEE104 pKa = 4.14IRR106 pKa = 11.84ATVAGSPFAIGQQQMAWYY124 pKa = 9.95YY125 pKa = 11.05DD126 pKa = 3.5ATHH129 pKa = 7.32DD130 pKa = 4.89SMFDD134 pKa = 3.21VMRR137 pKa = 11.84KK138 pKa = 9.02NKK140 pKa = 9.25YY141 pKa = 9.43ARR143 pKa = 11.84STMLHH148 pKa = 5.23TLIDD152 pKa = 4.14ASPSNDD158 pKa = 2.7AVLYY162 pKa = 10.21IPYY165 pKa = 9.99KK166 pKa = 9.72SYY168 pKa = 11.27RR169 pKa = 11.84SFLQTRR175 pKa = 11.84TTRR178 pKa = 11.84EE179 pKa = 4.03EE180 pKa = 3.62QGKK183 pKa = 8.63PLDD186 pKa = 4.74LGTLQIGVLNEE197 pKa = 3.8LLVPDD202 pKa = 5.12GATKK206 pKa = 10.1QVSLVLHH213 pKa = 5.98IRR215 pKa = 11.84FPNSEE220 pKa = 3.81FQGRR224 pKa = 11.84VSTALPGHH232 pKa = 6.95LL233 pKa = 4.33
MM1 pKa = 7.82SDD3 pKa = 3.27PHH5 pKa = 7.31GDD7 pKa = 3.49PAPVDD12 pKa = 3.38RR13 pKa = 11.84GGVISVEE20 pKa = 4.13VSEE23 pKa = 4.61EE24 pKa = 4.21SSGKK28 pKa = 9.5PDD30 pKa = 3.71FPTSIILSRR39 pKa = 11.84FCSVEE44 pKa = 3.77SEE46 pKa = 4.23QVYY49 pKa = 10.9KK50 pKa = 11.07DD51 pKa = 4.5LLNTWSILPPIIWEE65 pKa = 4.3GTTDD69 pKa = 4.98AVTTYY74 pKa = 10.53NLPYY78 pKa = 10.26YY79 pKa = 10.4AVEE82 pKa = 4.13SLKK85 pKa = 10.55DD86 pKa = 3.89YY87 pKa = 10.94PQMLPFRR94 pKa = 11.84MYY96 pKa = 10.23QYY98 pKa = 9.9WRR100 pKa = 11.84GDD102 pKa = 3.12IEE104 pKa = 4.14IRR106 pKa = 11.84ATVAGSPFAIGQQQMAWYY124 pKa = 9.95YY125 pKa = 11.05DD126 pKa = 3.5ATHH129 pKa = 7.32DD130 pKa = 4.89SMFDD134 pKa = 3.21VMRR137 pKa = 11.84KK138 pKa = 9.02NKK140 pKa = 9.25YY141 pKa = 9.43ARR143 pKa = 11.84STMLHH148 pKa = 5.23TLIDD152 pKa = 4.14ASPSNDD158 pKa = 2.7AVLYY162 pKa = 10.21IPYY165 pKa = 9.99KK166 pKa = 9.72SYY168 pKa = 11.27RR169 pKa = 11.84SFLQTRR175 pKa = 11.84TTRR178 pKa = 11.84EE179 pKa = 4.03EE180 pKa = 3.62QGKK183 pKa = 8.63PLDD186 pKa = 4.74LGTLQIGVLNEE197 pKa = 3.8LLVPDD202 pKa = 5.12GATKK206 pKa = 10.1QVSLVLHH213 pKa = 5.98IRR215 pKa = 11.84FPNSEE220 pKa = 3.81FQGRR224 pKa = 11.84VSTALPGHH232 pKa = 6.95LL233 pKa = 4.33
Molecular weight: 26.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJB6|A0A1L3KJB6_9VIRU SF3 helicase domain-containing protein OS=Sanxia water strider virus 7 OX=1923406 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84FNIPDD7 pKa = 3.9FGVVADD13 pKa = 4.61IFTTLMDD20 pKa = 4.53LLEE23 pKa = 4.8DD24 pKa = 4.16PKK26 pKa = 11.17TSIEE30 pKa = 4.25DD31 pKa = 3.4KK32 pKa = 11.15VKK34 pKa = 10.49IRR36 pKa = 11.84LDD38 pKa = 3.48LLSNEE43 pKa = 3.99GAYY46 pKa = 10.49GFDD49 pKa = 3.78LPSFGATRR57 pKa = 11.84RR58 pKa = 11.84VVKK61 pKa = 10.16EE62 pKa = 3.73VVPTIISEE70 pKa = 4.39DD71 pKa = 3.58KK72 pKa = 10.96LITPVKK78 pKa = 10.27RR79 pKa = 11.84GMSYY83 pKa = 11.51ANRR86 pKa = 11.84LQQRR90 pKa = 11.84VAKK93 pKa = 8.04PVKK96 pKa = 9.98YY97 pKa = 10.21IPPNCFCPEE106 pKa = 4.16CAPKK110 pKa = 10.35NAIYY114 pKa = 10.56SSSQYY119 pKa = 10.43YY120 pKa = 9.37RR121 pKa = 11.84EE122 pKa = 3.69LWIMNEE128 pKa = 3.2ARR130 pKa = 11.84KK131 pKa = 9.81RR132 pKa = 11.84GKK134 pKa = 9.86QLDD137 pKa = 3.9LKK139 pKa = 10.25KK140 pKa = 10.8ASSQATKK147 pKa = 10.81SRR149 pKa = 11.84TDD151 pKa = 3.56FVTITSKK158 pKa = 8.19THH160 pKa = 5.03ISARR164 pKa = 11.84NKK166 pKa = 10.19QILEE170 pKa = 4.02NVLLDD175 pKa = 3.6RR176 pKa = 11.84TSKK179 pKa = 9.02KK180 pKa = 7.35TKK182 pKa = 9.88KK183 pKa = 9.3MAVKK187 pKa = 10.21EE188 pKa = 4.28EE189 pKa = 4.11IVQSVSKK196 pKa = 9.73VTAQPKK202 pKa = 10.19KK203 pKa = 9.88KK204 pKa = 10.75VEE206 pKa = 4.15INSLPSSKK214 pKa = 10.55EE215 pKa = 3.54EE216 pKa = 3.96KK217 pKa = 9.55TNSLVFVSKK226 pKa = 10.75RR227 pKa = 11.84KK228 pKa = 6.08QTRR231 pKa = 11.84NLTSFSRR238 pKa = 11.84SDD240 pKa = 3.27EE241 pKa = 4.19KK242 pKa = 10.88QSRR245 pKa = 11.84IYY247 pKa = 10.39TPIIDD252 pKa = 4.08PNYY255 pKa = 8.89KK256 pKa = 9.95PRR258 pKa = 11.84IKK260 pKa = 10.48VEE262 pKa = 3.5KK263 pKa = 10.43SGFIKK268 pKa = 10.67SRR270 pKa = 11.84TVNNAKK276 pKa = 8.79TVEE279 pKa = 4.07ITPFLAKK286 pKa = 10.48LPPQVVNSTSEE297 pKa = 4.13TVSVKK302 pKa = 10.19KK303 pKa = 9.89VQPEE307 pKa = 4.19PEE309 pKa = 4.35TPKK312 pKa = 10.57QPNNRR317 pKa = 11.84IRR319 pKa = 11.84TKK321 pKa = 10.51LIRR324 pKa = 11.84EE325 pKa = 3.82FRR327 pKa = 11.84EE328 pKa = 4.19LIRR331 pKa = 11.84VSGLKK336 pKa = 9.25GTFSKK341 pKa = 10.6EE342 pKa = 3.77GKK344 pKa = 6.29PTKK347 pKa = 10.01DD348 pKa = 2.8TVLVLDD354 pKa = 4.68ALISDD359 pKa = 3.98MRR361 pKa = 11.84IKK363 pKa = 10.77ADD365 pKa = 3.4QLKK368 pKa = 10.48VSPEE372 pKa = 4.19SINLKK377 pKa = 8.95STLQSTLGTAFFVQSLNVV395 pKa = 3.25
MM1 pKa = 7.9RR2 pKa = 11.84FNIPDD7 pKa = 3.9FGVVADD13 pKa = 4.61IFTTLMDD20 pKa = 4.53LLEE23 pKa = 4.8DD24 pKa = 4.16PKK26 pKa = 11.17TSIEE30 pKa = 4.25DD31 pKa = 3.4KK32 pKa = 11.15VKK34 pKa = 10.49IRR36 pKa = 11.84LDD38 pKa = 3.48LLSNEE43 pKa = 3.99GAYY46 pKa = 10.49GFDD49 pKa = 3.78LPSFGATRR57 pKa = 11.84RR58 pKa = 11.84VVKK61 pKa = 10.16EE62 pKa = 3.73VVPTIISEE70 pKa = 4.39DD71 pKa = 3.58KK72 pKa = 10.96LITPVKK78 pKa = 10.27RR79 pKa = 11.84GMSYY83 pKa = 11.51ANRR86 pKa = 11.84LQQRR90 pKa = 11.84VAKK93 pKa = 8.04PVKK96 pKa = 9.98YY97 pKa = 10.21IPPNCFCPEE106 pKa = 4.16CAPKK110 pKa = 10.35NAIYY114 pKa = 10.56SSSQYY119 pKa = 10.43YY120 pKa = 9.37RR121 pKa = 11.84EE122 pKa = 3.69LWIMNEE128 pKa = 3.2ARR130 pKa = 11.84KK131 pKa = 9.81RR132 pKa = 11.84GKK134 pKa = 9.86QLDD137 pKa = 3.9LKK139 pKa = 10.25KK140 pKa = 10.8ASSQATKK147 pKa = 10.81SRR149 pKa = 11.84TDD151 pKa = 3.56FVTITSKK158 pKa = 8.19THH160 pKa = 5.03ISARR164 pKa = 11.84NKK166 pKa = 10.19QILEE170 pKa = 4.02NVLLDD175 pKa = 3.6RR176 pKa = 11.84TSKK179 pKa = 9.02KK180 pKa = 7.35TKK182 pKa = 9.88KK183 pKa = 9.3MAVKK187 pKa = 10.21EE188 pKa = 4.28EE189 pKa = 4.11IVQSVSKK196 pKa = 9.73VTAQPKK202 pKa = 10.19KK203 pKa = 9.88KK204 pKa = 10.75VEE206 pKa = 4.15INSLPSSKK214 pKa = 10.55EE215 pKa = 3.54EE216 pKa = 3.96KK217 pKa = 9.55TNSLVFVSKK226 pKa = 10.75RR227 pKa = 11.84KK228 pKa = 6.08QTRR231 pKa = 11.84NLTSFSRR238 pKa = 11.84SDD240 pKa = 3.27EE241 pKa = 4.19KK242 pKa = 10.88QSRR245 pKa = 11.84IYY247 pKa = 10.39TPIIDD252 pKa = 4.08PNYY255 pKa = 8.89KK256 pKa = 9.95PRR258 pKa = 11.84IKK260 pKa = 10.48VEE262 pKa = 3.5KK263 pKa = 10.43SGFIKK268 pKa = 10.67SRR270 pKa = 11.84TVNNAKK276 pKa = 8.79TVEE279 pKa = 4.07ITPFLAKK286 pKa = 10.48LPPQVVNSTSEE297 pKa = 4.13TVSVKK302 pKa = 10.19KK303 pKa = 9.89VQPEE307 pKa = 4.19PEE309 pKa = 4.35TPKK312 pKa = 10.57QPNNRR317 pKa = 11.84IRR319 pKa = 11.84TKK321 pKa = 10.51LIRR324 pKa = 11.84EE325 pKa = 3.82FRR327 pKa = 11.84EE328 pKa = 4.19LIRR331 pKa = 11.84VSGLKK336 pKa = 9.25GTFSKK341 pKa = 10.6EE342 pKa = 3.77GKK344 pKa = 6.29PTKK347 pKa = 10.01DD348 pKa = 2.8TVLVLDD354 pKa = 4.68ALISDD359 pKa = 3.98MRR361 pKa = 11.84IKK363 pKa = 10.77ADD365 pKa = 3.4QLKK368 pKa = 10.48VSPEE372 pKa = 4.19SINLKK377 pKa = 8.95STLQSTLGTAFFVQSLNVV395 pKa = 3.25
Molecular weight: 44.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3033 |
233 |
2405 |
1011.0 |
114.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.341 ± 0.255 | 2.275 ± 0.815 |
5.935 ± 0.53 | 5.308 ± 0.254 |
5.44 ± 0.884 | 4.946 ± 0.693 |
1.945 ± 0.569 | 6.924 ± 0.344 |
6.034 ± 1.884 | 7.946 ± 0.252 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.143 ± 0.238 | 4.286 ± 0.39 |
5.671 ± 0.472 | 3.825 ± 0.261 |
5.176 ± 0.397 | 9.034 ± 0.201 |
5.869 ± 0.812 | 6.462 ± 0.7 |
1.352 ± 0.356 | 4.055 ± 0.659 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
