
Sorghum chlorotic spot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Furovirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
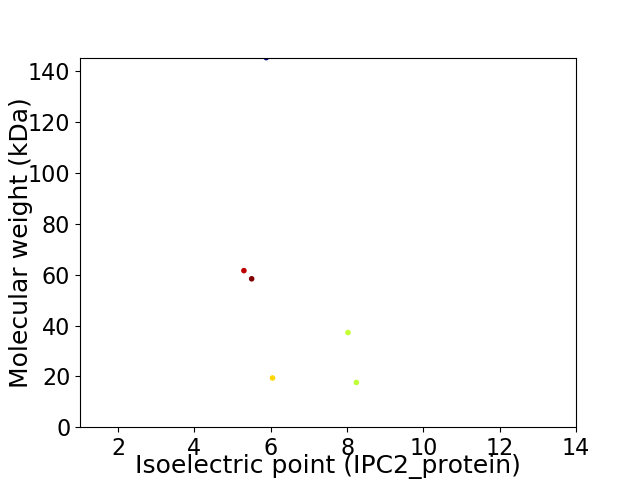
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9JGJ4|Q9JGJ4_9VIRU Capsid protein OS=Sorghum chlorotic spot virus OX=107804 GN=CP PE=4 SV=1
WW1 pKa = 7.68GGVLSASRR9 pKa = 11.84EE10 pKa = 4.39SVVEE14 pKa = 3.77NAVLRR19 pKa = 11.84RR20 pKa = 11.84LTAKK24 pKa = 10.61DD25 pKa = 3.52LGLLPRR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.71TNGTVLQDD41 pKa = 3.89GPDD44 pKa = 3.07ILYY47 pKa = 10.82EE48 pKa = 4.04EE49 pKa = 5.52LEE51 pKa = 4.46DD52 pKa = 5.67ALLASNTSVYY62 pKa = 10.31NPHH65 pKa = 6.78AAIEE69 pKa = 4.31EE70 pKa = 4.39YY71 pKa = 10.57EE72 pKa = 4.05IDD74 pKa = 3.73TGDD77 pKa = 3.87GEE79 pKa = 4.41QLRR82 pKa = 11.84HH83 pKa = 5.35EE84 pKa = 4.6VSHH87 pKa = 6.61TEE89 pKa = 3.86DD90 pKa = 2.8SDD92 pKa = 3.76YY93 pKa = 11.18RR94 pKa = 11.84ILYY97 pKa = 9.91LFLILCAILMTLAMMLSRR115 pKa = 11.84AFVNFLYY122 pKa = 10.43NRR124 pKa = 11.84RR125 pKa = 11.84DD126 pKa = 3.05HH127 pKa = 6.9RR128 pKa = 11.84RR129 pKa = 11.84FGGPCEE135 pKa = 3.82RR136 pKa = 11.84CARR139 pKa = 11.84RR140 pKa = 11.84LWRR143 pKa = 11.84FRR145 pKa = 11.84LWLVGLAMRR154 pKa = 11.84RR155 pKa = 11.84YY156 pKa = 10.59GDD158 pKa = 3.45LQVTRR163 pKa = 11.84KK164 pKa = 9.02RR165 pKa = 11.84HH166 pKa = 4.18TRR168 pKa = 11.84SAMRR172 pKa = 11.84QYY174 pKa = 11.12LNSYY178 pKa = 9.58EE179 pKa = 4.28DD180 pKa = 4.33QTDD183 pKa = 2.98LWQSYY188 pKa = 10.17EE189 pKa = 3.93GVTPEE194 pKa = 4.02EE195 pKa = 4.04ALIVADD201 pKa = 3.13QHH203 pKa = 6.71LFVDD207 pKa = 5.18AVDD210 pKa = 3.85QLASGGGDD218 pKa = 3.48LQCVRR223 pKa = 11.84TAQTISEE230 pKa = 4.43RR231 pKa = 11.84YY232 pKa = 8.3GVKK235 pKa = 10.49NPALQDD241 pKa = 3.54KK242 pKa = 10.32LDD244 pKa = 3.68EE245 pKa = 4.33FARR248 pKa = 11.84RR249 pKa = 11.84LAIQVRR255 pKa = 11.84FEE257 pKa = 4.53DD258 pKa = 3.73TDD260 pKa = 3.56ATPGFLDD267 pKa = 3.96LRR269 pKa = 11.84NASMQEE275 pKa = 3.89VLDD278 pKa = 4.64CDD280 pKa = 5.15LSEE283 pKa = 4.12QTINPQYY290 pKa = 11.04NQICALQHH298 pKa = 4.7MFAMVGKK305 pKa = 8.92PAFTDD310 pKa = 3.87FVIAYY315 pKa = 8.45KK316 pKa = 9.35GTYY319 pKa = 7.8YY320 pKa = 10.31PCVIEE325 pKa = 4.26AQRR328 pKa = 11.84KK329 pKa = 6.59HH330 pKa = 8.2AEE332 pKa = 4.08MLGTDD337 pKa = 4.82PDD339 pKa = 4.6DD340 pKa = 3.16KK341 pKa = 10.62WGPLEE346 pKa = 3.9WMYY349 pKa = 10.95CVIMVGSLASFGFATVKK366 pKa = 10.9GIVKK370 pKa = 10.03LSQMVKK376 pKa = 8.56NWLRR380 pKa = 11.84LRR382 pKa = 11.84ALRR385 pKa = 11.84ALPSVTHH392 pKa = 6.5GGDD395 pKa = 3.36GGDD398 pKa = 3.26GGLPFFEE405 pKa = 5.04MEE407 pKa = 4.73DD408 pKa = 3.31FTARR412 pKa = 11.84LRR414 pKa = 11.84VLNEE418 pKa = 3.58FVANMGEE425 pKa = 4.28QVVDD429 pKa = 3.46TSVEE433 pKa = 3.91DD434 pKa = 3.56LRR436 pKa = 11.84RR437 pKa = 11.84DD438 pKa = 3.52LVSVLTAHH446 pKa = 6.8SGSEE450 pKa = 4.03PPAVDD455 pKa = 3.15RR456 pKa = 11.84PYY458 pKa = 11.33LEE460 pKa = 4.21GVGIGRR466 pKa = 11.84EE467 pKa = 4.09VDD469 pKa = 3.5EE470 pKa = 5.57LPASTEE476 pKa = 3.78IEE478 pKa = 4.33GEE480 pKa = 4.05QGEE483 pKa = 4.82SLFDD487 pKa = 3.93PGLRR491 pKa = 11.84IKK493 pKa = 10.5RR494 pKa = 11.84WEE496 pKa = 4.05TEE498 pKa = 3.55AAVPKK503 pKa = 10.59GVGGFSSFIRR513 pKa = 11.84TSGSVSGRR521 pKa = 11.84ARR523 pKa = 11.84MSAQNSVSRR532 pKa = 11.84VKK534 pKa = 10.68LRR536 pKa = 11.84NASMMKK542 pKa = 10.05SKK544 pKa = 11.02RR545 pKa = 11.84LMAAA549 pKa = 3.44
WW1 pKa = 7.68GGVLSASRR9 pKa = 11.84EE10 pKa = 4.39SVVEE14 pKa = 3.77NAVLRR19 pKa = 11.84RR20 pKa = 11.84LTAKK24 pKa = 10.61DD25 pKa = 3.52LGLLPRR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.71TNGTVLQDD41 pKa = 3.89GPDD44 pKa = 3.07ILYY47 pKa = 10.82EE48 pKa = 4.04EE49 pKa = 5.52LEE51 pKa = 4.46DD52 pKa = 5.67ALLASNTSVYY62 pKa = 10.31NPHH65 pKa = 6.78AAIEE69 pKa = 4.31EE70 pKa = 4.39YY71 pKa = 10.57EE72 pKa = 4.05IDD74 pKa = 3.73TGDD77 pKa = 3.87GEE79 pKa = 4.41QLRR82 pKa = 11.84HH83 pKa = 5.35EE84 pKa = 4.6VSHH87 pKa = 6.61TEE89 pKa = 3.86DD90 pKa = 2.8SDD92 pKa = 3.76YY93 pKa = 11.18RR94 pKa = 11.84ILYY97 pKa = 9.91LFLILCAILMTLAMMLSRR115 pKa = 11.84AFVNFLYY122 pKa = 10.43NRR124 pKa = 11.84RR125 pKa = 11.84DD126 pKa = 3.05HH127 pKa = 6.9RR128 pKa = 11.84RR129 pKa = 11.84FGGPCEE135 pKa = 3.82RR136 pKa = 11.84CARR139 pKa = 11.84RR140 pKa = 11.84LWRR143 pKa = 11.84FRR145 pKa = 11.84LWLVGLAMRR154 pKa = 11.84RR155 pKa = 11.84YY156 pKa = 10.59GDD158 pKa = 3.45LQVTRR163 pKa = 11.84KK164 pKa = 9.02RR165 pKa = 11.84HH166 pKa = 4.18TRR168 pKa = 11.84SAMRR172 pKa = 11.84QYY174 pKa = 11.12LNSYY178 pKa = 9.58EE179 pKa = 4.28DD180 pKa = 4.33QTDD183 pKa = 2.98LWQSYY188 pKa = 10.17EE189 pKa = 3.93GVTPEE194 pKa = 4.02EE195 pKa = 4.04ALIVADD201 pKa = 3.13QHH203 pKa = 6.71LFVDD207 pKa = 5.18AVDD210 pKa = 3.85QLASGGGDD218 pKa = 3.48LQCVRR223 pKa = 11.84TAQTISEE230 pKa = 4.43RR231 pKa = 11.84YY232 pKa = 8.3GVKK235 pKa = 10.49NPALQDD241 pKa = 3.54KK242 pKa = 10.32LDD244 pKa = 3.68EE245 pKa = 4.33FARR248 pKa = 11.84RR249 pKa = 11.84LAIQVRR255 pKa = 11.84FEE257 pKa = 4.53DD258 pKa = 3.73TDD260 pKa = 3.56ATPGFLDD267 pKa = 3.96LRR269 pKa = 11.84NASMQEE275 pKa = 3.89VLDD278 pKa = 4.64CDD280 pKa = 5.15LSEE283 pKa = 4.12QTINPQYY290 pKa = 11.04NQICALQHH298 pKa = 4.7MFAMVGKK305 pKa = 8.92PAFTDD310 pKa = 3.87FVIAYY315 pKa = 8.45KK316 pKa = 9.35GTYY319 pKa = 7.8YY320 pKa = 10.31PCVIEE325 pKa = 4.26AQRR328 pKa = 11.84KK329 pKa = 6.59HH330 pKa = 8.2AEE332 pKa = 4.08MLGTDD337 pKa = 4.82PDD339 pKa = 4.6DD340 pKa = 3.16KK341 pKa = 10.62WGPLEE346 pKa = 3.9WMYY349 pKa = 10.95CVIMVGSLASFGFATVKK366 pKa = 10.9GIVKK370 pKa = 10.03LSQMVKK376 pKa = 8.56NWLRR380 pKa = 11.84LRR382 pKa = 11.84ALRR385 pKa = 11.84ALPSVTHH392 pKa = 6.5GGDD395 pKa = 3.36GGDD398 pKa = 3.26GGLPFFEE405 pKa = 5.04MEE407 pKa = 4.73DD408 pKa = 3.31FTARR412 pKa = 11.84LRR414 pKa = 11.84VLNEE418 pKa = 3.58FVANMGEE425 pKa = 4.28QVVDD429 pKa = 3.46TSVEE433 pKa = 3.91DD434 pKa = 3.56LRR436 pKa = 11.84RR437 pKa = 11.84DD438 pKa = 3.52LVSVLTAHH446 pKa = 6.8SGSEE450 pKa = 4.03PPAVDD455 pKa = 3.15RR456 pKa = 11.84PYY458 pKa = 11.33LEE460 pKa = 4.21GVGIGRR466 pKa = 11.84EE467 pKa = 4.09VDD469 pKa = 3.5EE470 pKa = 5.57LPASTEE476 pKa = 3.78IEE478 pKa = 4.33GEE480 pKa = 4.05QGEE483 pKa = 4.82SLFDD487 pKa = 3.93PGLRR491 pKa = 11.84IKK493 pKa = 10.5RR494 pKa = 11.84WEE496 pKa = 4.05TEE498 pKa = 3.55AAVPKK503 pKa = 10.59GVGGFSSFIRR513 pKa = 11.84TSGSVSGRR521 pKa = 11.84ARR523 pKa = 11.84MSAQNSVSRR532 pKa = 11.84VKK534 pKa = 10.68LRR536 pKa = 11.84NASMMKK542 pKa = 10.05SKK544 pKa = 11.02RR545 pKa = 11.84LMAAA549 pKa = 3.44
Molecular weight: 61.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9JGJ3|Q9JGJ3_9VIRU 62-kDa readthrough protein (Fragment) OS=Sorghum chlorotic spot virus OX=107804 GN=RT PE=4 SV=1
MM1 pKa = 6.92TVSTIHH7 pKa = 5.49SCEE10 pKa = 3.67RR11 pKa = 11.84CLEE14 pKa = 4.04GRR16 pKa = 11.84TSLRR20 pKa = 11.84CEE22 pKa = 3.49NKK24 pKa = 9.57YY25 pKa = 10.58RR26 pKa = 11.84LSVYY30 pKa = 8.61QSRR33 pKa = 11.84QVEE36 pKa = 4.23KK37 pKa = 10.47SAYY40 pKa = 8.57ACKK43 pKa = 10.01ISQFGVPCGMPAQFEE58 pKa = 4.62LDD60 pKa = 3.6GEE62 pKa = 4.55TLKK65 pKa = 10.94VVCDD69 pKa = 4.97GYY71 pKa = 11.48CGLKK75 pKa = 9.3HH76 pKa = 6.77KK77 pKa = 11.04NMAEE81 pKa = 3.99SGSWRR86 pKa = 11.84GTLLVILQKK95 pKa = 10.55EE96 pKa = 4.27LEE98 pKa = 4.09ALQLKK103 pKa = 9.76EE104 pKa = 4.2EE105 pKa = 4.05QLKK108 pKa = 8.82TRR110 pKa = 11.84IAEE113 pKa = 4.0VTQQHH118 pKa = 6.71DD119 pKa = 3.73LVMAEE124 pKa = 4.07TAAVLRR130 pKa = 11.84PDD132 pKa = 4.28SPPKK136 pKa = 11.11AMVTTNSRR144 pKa = 11.84VKK146 pKa = 9.96YY147 pKa = 9.86VRR149 pKa = 11.84RR150 pKa = 11.84KK151 pKa = 8.03PAPRR155 pKa = 11.84MM156 pKa = 3.56
MM1 pKa = 6.92TVSTIHH7 pKa = 5.49SCEE10 pKa = 3.67RR11 pKa = 11.84CLEE14 pKa = 4.04GRR16 pKa = 11.84TSLRR20 pKa = 11.84CEE22 pKa = 3.49NKK24 pKa = 9.57YY25 pKa = 10.58RR26 pKa = 11.84LSVYY30 pKa = 8.61QSRR33 pKa = 11.84QVEE36 pKa = 4.23KK37 pKa = 10.47SAYY40 pKa = 8.57ACKK43 pKa = 10.01ISQFGVPCGMPAQFEE58 pKa = 4.62LDD60 pKa = 3.6GEE62 pKa = 4.55TLKK65 pKa = 10.94VVCDD69 pKa = 4.97GYY71 pKa = 11.48CGLKK75 pKa = 9.3HH76 pKa = 6.77KK77 pKa = 11.04NMAEE81 pKa = 3.99SGSWRR86 pKa = 11.84GTLLVILQKK95 pKa = 10.55EE96 pKa = 4.27LEE98 pKa = 4.09ALQLKK103 pKa = 9.76EE104 pKa = 4.2EE105 pKa = 4.05QLKK108 pKa = 8.82TRR110 pKa = 11.84IAEE113 pKa = 4.0VTQQHH118 pKa = 6.71DD119 pKa = 3.73LVMAEE124 pKa = 4.07TAAVLRR130 pKa = 11.84PDD132 pKa = 4.28SPPKK136 pKa = 11.11AMVTTNSRR144 pKa = 11.84VKK146 pKa = 9.96YY147 pKa = 9.86VRR149 pKa = 11.84RR150 pKa = 11.84KK151 pKa = 8.03PAPRR155 pKa = 11.84MM156 pKa = 3.56
Molecular weight: 17.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2991 |
156 |
1277 |
498.5 |
56.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.954 ± 0.806 | 1.906 ± 0.314 |
6.653 ± 0.361 | 7.322 ± 0.213 |
4.313 ± 0.547 | 5.583 ± 0.563 |
1.705 ± 0.249 | 4.447 ± 0.5 |
6.687 ± 0.922 | 8.76 ± 0.6 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.976 ± 0.404 | 4.346 ± 0.402 |
3.243 ± 0.298 | 3.644 ± 0.32 |
6.553 ± 0.504 | 6.653 ± 0.324 |
5.517 ± 0.483 | 8.124 ± 0.569 |
1.17 ± 0.12 | 3.444 ± 0.258 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
