
Murine coronavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Embecovirus
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
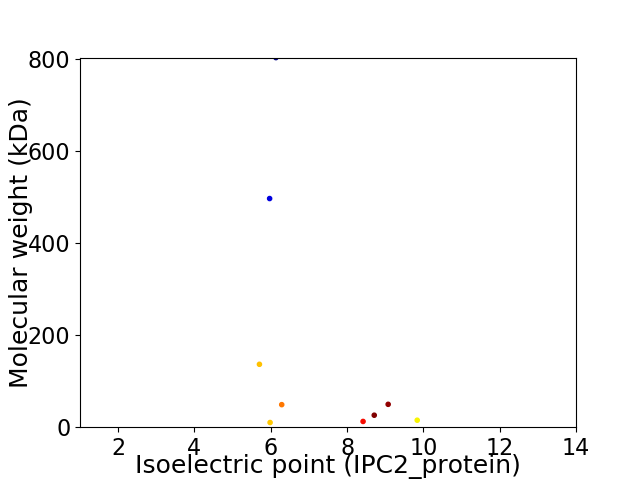
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I7B990|I7B990_9BETC Nucleoprotein OS=Murine coronavirus OX=694005 GN=N PE=3 SV=1
MM1 pKa = 7.47LFVFILLLPSCLGYY15 pKa = 10.36IGDD18 pKa = 4.3FRR20 pKa = 11.84CIQTVNYY27 pKa = 9.86NGNNASAPSISTEE40 pKa = 3.73AVDD43 pKa = 3.61VSKK46 pKa = 11.15GRR48 pKa = 11.84GTYY51 pKa = 9.22YY52 pKa = 11.01VLDD55 pKa = 3.67RR56 pKa = 11.84VYY58 pKa = 11.47LNATLLLTGYY68 pKa = 10.58YY69 pKa = 9.74PVDD72 pKa = 3.26GSNYY76 pKa = 9.77RR77 pKa = 11.84NLALTGTNTLSLTWFKK93 pKa = 11.13PPFLSEE99 pKa = 4.04FNDD102 pKa = 5.06GIFAKK107 pKa = 10.4VQNLKK112 pKa = 9.78TNTPTGATSYY122 pKa = 10.66FPTIVIGSLFGNTSYY137 pKa = 10.68TVVLEE142 pKa = 4.17PYY144 pKa = 10.14NNIIMASVCTYY155 pKa = 7.55TICQLPYY162 pKa = 10.03TPCKK166 pKa = 10.23PNTNGNRR173 pKa = 11.84VIGFWHH179 pKa = 7.05TDD181 pKa = 3.24VKK183 pKa = 11.12PPICLLKK190 pKa = 10.88RR191 pKa = 11.84NFTFNVNAPWLYY203 pKa = 10.39FHH205 pKa = 7.79FYY207 pKa = 8.91QQGGTFYY214 pKa = 10.77AYY216 pKa = 10.83YY217 pKa = 10.42ADD219 pKa = 4.72KK220 pKa = 10.83PSATTFLFSVYY231 pKa = 10.07IGDD234 pKa = 3.89ILTQYY239 pKa = 10.48FVLPFICTPTAGSTLAPLYY258 pKa = 9.98WVTPLLKK265 pKa = 10.15RR266 pKa = 11.84QYY268 pKa = 10.93LFNFNEE274 pKa = 4.21KK275 pKa = 10.49GVITSAVDD283 pKa = 3.33CASSYY288 pKa = 10.66ISEE291 pKa = 4.54IKK293 pKa = 10.69CKK295 pKa = 8.53TQSLLPSTGVYY306 pKa = 10.24DD307 pKa = 3.93LSGYY311 pKa = 6.78TVQPVGVVYY320 pKa = 10.46RR321 pKa = 11.84RR322 pKa = 11.84VPNLPDD328 pKa = 3.5CKK330 pKa = 9.93IEE332 pKa = 3.81EE333 pKa = 4.32WLTAKK338 pKa = 10.27SVPSPLNWEE347 pKa = 3.58RR348 pKa = 11.84RR349 pKa = 11.84TFQNCNFNLSSLLRR363 pKa = 11.84YY364 pKa = 8.69VQAEE368 pKa = 4.13SLSCNNIDD376 pKa = 3.31ASKK379 pKa = 10.66VYY381 pKa = 9.95GMCFGSVSVDD391 pKa = 2.67KK392 pKa = 10.62FAIPRR397 pKa = 11.84SRR399 pKa = 11.84QIDD402 pKa = 3.88LQIGNSGFLQTANYY416 pKa = 9.74KK417 pKa = 9.91IDD419 pKa = 3.64TAATSCQLYY428 pKa = 10.65YY429 pKa = 10.87SLPKK433 pKa = 10.92NNVTINNYY441 pKa = 9.54NPSSWNRR448 pKa = 11.84RR449 pKa = 11.84YY450 pKa = 10.28GFKK453 pKa = 11.13VNDD456 pKa = 3.59RR457 pKa = 11.84CQIFANILLNGINSGTTCSTDD478 pKa = 3.14LQLPNTEE485 pKa = 4.21VATGVCVRR493 pKa = 11.84YY494 pKa = 9.74DD495 pKa = 3.44LYY497 pKa = 11.29GITGQGVFKK506 pKa = 10.25EE507 pKa = 4.64VKK509 pKa = 9.62ADD511 pKa = 4.25YY512 pKa = 10.05YY513 pKa = 11.52NSWQALLYY521 pKa = 10.55DD522 pKa = 4.03VNGNLNGFRR531 pKa = 11.84DD532 pKa = 3.65LTTNKK537 pKa = 9.29TYY539 pKa = 10.09TIRR542 pKa = 11.84SCYY545 pKa = 9.26SGRR548 pKa = 11.84VSAAYY553 pKa = 9.67HH554 pKa = 6.17KK555 pKa = 8.08EE556 pKa = 3.96APEE559 pKa = 3.78PALLYY564 pKa = 10.9RR565 pKa = 11.84NINCSYY571 pKa = 10.95VFTNNISRR579 pKa = 11.84EE580 pKa = 4.24EE581 pKa = 3.99NPLNYY586 pKa = 9.38FDD588 pKa = 5.71SYY590 pKa = 10.97LGCVVNADD598 pKa = 3.38NRR600 pKa = 11.84TDD602 pKa = 3.5EE603 pKa = 5.07ALPNCDD609 pKa = 3.43LRR611 pKa = 11.84MGAGLCVDD619 pKa = 3.67YY620 pKa = 11.11SKK622 pKa = 11.14SRR624 pKa = 11.84RR625 pKa = 11.84ARR627 pKa = 11.84RR628 pKa = 11.84SVSTGYY634 pKa = 11.01RR635 pKa = 11.84LTTFEE640 pKa = 5.37PYY642 pKa = 9.99MPMLVNDD649 pKa = 4.28SVQSVGGLYY658 pKa = 10.45EE659 pKa = 3.97MQIPTNFTIGHH670 pKa = 6.13HH671 pKa = 5.77EE672 pKa = 4.09EE673 pKa = 4.92FIQIRR678 pKa = 11.84APKK681 pKa = 8.75VTIDD685 pKa = 3.67CAAFVCGDD693 pKa = 3.4NAACRR698 pKa = 11.84QQLVEE703 pKa = 4.55YY704 pKa = 10.87GSFCDD709 pKa = 4.0NVNAILNEE717 pKa = 3.97VNNLLDD723 pKa = 4.08NMQLQVASALMQGVTISSRR742 pKa = 11.84LPDD745 pKa = 5.17GISGPIDD752 pKa = 4.86DD753 pKa = 5.34INFSPLLGCIGSTCAEE769 pKa = 4.69DD770 pKa = 4.47GNGPSAIRR778 pKa = 11.84GRR780 pKa = 11.84SAIEE784 pKa = 4.11DD785 pKa = 3.76LLFDD789 pKa = 4.06KK790 pKa = 11.25VKK792 pKa = 10.82LSDD795 pKa = 3.34VGFVEE800 pKa = 6.19AYY802 pKa = 9.87NNCTGGQEE810 pKa = 4.32VRR812 pKa = 11.84DD813 pKa = 3.97LLCVQSFNGIKK824 pKa = 10.05VLPPVLSEE832 pKa = 4.18SQISGYY838 pKa = 7.58TAGATAAAMFPPWTAAAGVPFSLNVQYY865 pKa = 10.59RR866 pKa = 11.84INGLGVTMNVLSEE879 pKa = 4.11NQKK882 pKa = 10.06MIASAFNNALGAIQEE897 pKa = 4.62GFDD900 pKa = 3.73ATNSALGKK908 pKa = 9.37IQSVVNANAEE918 pKa = 4.13ALNNLLNQLSNRR930 pKa = 11.84FGAISASLQEE940 pKa = 4.0ILTRR944 pKa = 11.84LDD946 pKa = 3.15AVEE949 pKa = 5.13AKK951 pKa = 10.57AQIDD955 pKa = 3.56RR956 pKa = 11.84LINGRR961 pKa = 11.84LTALNAYY968 pKa = 9.21ISKK971 pKa = 9.99QLSDD975 pKa = 3.23STLIKK980 pKa = 10.46FSAAQAIEE988 pKa = 4.11KK989 pKa = 10.61VNEE992 pKa = 4.28CVKK995 pKa = 10.93SQTTRR1000 pKa = 11.84INFCGNGNHH1009 pKa = 6.86ILSLVQNAPYY1019 pKa = 9.9GLCFIHH1025 pKa = 6.84FSYY1028 pKa = 11.19VPTSFKK1034 pKa = 9.67TANVSPGLCISGDD1047 pKa = 3.15RR1048 pKa = 11.84GLAPKK1053 pKa = 10.3AGYY1056 pKa = 8.93FVQDD1060 pKa = 3.0NGEE1063 pKa = 4.07WKK1065 pKa = 8.91FTGSNYY1071 pKa = 10.17YY1072 pKa = 9.98YY1073 pKa = 9.83PEE1075 pKa = 5.68PITDD1079 pKa = 3.59KK1080 pKa = 10.85NSVVMISCAVNYY1092 pKa = 7.8TKK1094 pKa = 10.77APEE1097 pKa = 4.02VFLNNSIPNLPDD1109 pKa = 3.66FKK1111 pKa = 11.37EE1112 pKa = 4.4EE1113 pKa = 3.64LDD1115 pKa = 3.46KK1116 pKa = 11.03WFKK1119 pKa = 10.55NQTSIAPDD1127 pKa = 3.69LSLDD1131 pKa = 3.81FEE1133 pKa = 4.58KK1134 pKa = 11.4LNVTFLDD1141 pKa = 3.44LTYY1144 pKa = 11.03EE1145 pKa = 4.04MNRR1148 pKa = 11.84IQDD1151 pKa = 4.14AIKK1154 pKa = 10.34KK1155 pKa = 10.05LNEE1158 pKa = 3.82SYY1160 pKa = 11.14INLKK1164 pKa = 10.23EE1165 pKa = 3.97VGTYY1169 pKa = 9.32EE1170 pKa = 5.21MYY1172 pKa = 11.04VKK1174 pKa = 9.96WPWYY1178 pKa = 8.24VWLLIGLAGVAVCVLLFFICCCTGCGSCCFRR1209 pKa = 11.84KK1210 pKa = 10.28CGSCCDD1216 pKa = 3.87EE1217 pKa = 4.38YY1218 pKa = 11.48GGHH1221 pKa = 6.11QDD1223 pKa = 4.52SIVIHH1228 pKa = 6.54NISAHH1233 pKa = 5.82EE1234 pKa = 4.06DD1235 pKa = 3.09
MM1 pKa = 7.47LFVFILLLPSCLGYY15 pKa = 10.36IGDD18 pKa = 4.3FRR20 pKa = 11.84CIQTVNYY27 pKa = 9.86NGNNASAPSISTEE40 pKa = 3.73AVDD43 pKa = 3.61VSKK46 pKa = 11.15GRR48 pKa = 11.84GTYY51 pKa = 9.22YY52 pKa = 11.01VLDD55 pKa = 3.67RR56 pKa = 11.84VYY58 pKa = 11.47LNATLLLTGYY68 pKa = 10.58YY69 pKa = 9.74PVDD72 pKa = 3.26GSNYY76 pKa = 9.77RR77 pKa = 11.84NLALTGTNTLSLTWFKK93 pKa = 11.13PPFLSEE99 pKa = 4.04FNDD102 pKa = 5.06GIFAKK107 pKa = 10.4VQNLKK112 pKa = 9.78TNTPTGATSYY122 pKa = 10.66FPTIVIGSLFGNTSYY137 pKa = 10.68TVVLEE142 pKa = 4.17PYY144 pKa = 10.14NNIIMASVCTYY155 pKa = 7.55TICQLPYY162 pKa = 10.03TPCKK166 pKa = 10.23PNTNGNRR173 pKa = 11.84VIGFWHH179 pKa = 7.05TDD181 pKa = 3.24VKK183 pKa = 11.12PPICLLKK190 pKa = 10.88RR191 pKa = 11.84NFTFNVNAPWLYY203 pKa = 10.39FHH205 pKa = 7.79FYY207 pKa = 8.91QQGGTFYY214 pKa = 10.77AYY216 pKa = 10.83YY217 pKa = 10.42ADD219 pKa = 4.72KK220 pKa = 10.83PSATTFLFSVYY231 pKa = 10.07IGDD234 pKa = 3.89ILTQYY239 pKa = 10.48FVLPFICTPTAGSTLAPLYY258 pKa = 9.98WVTPLLKK265 pKa = 10.15RR266 pKa = 11.84QYY268 pKa = 10.93LFNFNEE274 pKa = 4.21KK275 pKa = 10.49GVITSAVDD283 pKa = 3.33CASSYY288 pKa = 10.66ISEE291 pKa = 4.54IKK293 pKa = 10.69CKK295 pKa = 8.53TQSLLPSTGVYY306 pKa = 10.24DD307 pKa = 3.93LSGYY311 pKa = 6.78TVQPVGVVYY320 pKa = 10.46RR321 pKa = 11.84RR322 pKa = 11.84VPNLPDD328 pKa = 3.5CKK330 pKa = 9.93IEE332 pKa = 3.81EE333 pKa = 4.32WLTAKK338 pKa = 10.27SVPSPLNWEE347 pKa = 3.58RR348 pKa = 11.84RR349 pKa = 11.84TFQNCNFNLSSLLRR363 pKa = 11.84YY364 pKa = 8.69VQAEE368 pKa = 4.13SLSCNNIDD376 pKa = 3.31ASKK379 pKa = 10.66VYY381 pKa = 9.95GMCFGSVSVDD391 pKa = 2.67KK392 pKa = 10.62FAIPRR397 pKa = 11.84SRR399 pKa = 11.84QIDD402 pKa = 3.88LQIGNSGFLQTANYY416 pKa = 9.74KK417 pKa = 9.91IDD419 pKa = 3.64TAATSCQLYY428 pKa = 10.65YY429 pKa = 10.87SLPKK433 pKa = 10.92NNVTINNYY441 pKa = 9.54NPSSWNRR448 pKa = 11.84RR449 pKa = 11.84YY450 pKa = 10.28GFKK453 pKa = 11.13VNDD456 pKa = 3.59RR457 pKa = 11.84CQIFANILLNGINSGTTCSTDD478 pKa = 3.14LQLPNTEE485 pKa = 4.21VATGVCVRR493 pKa = 11.84YY494 pKa = 9.74DD495 pKa = 3.44LYY497 pKa = 11.29GITGQGVFKK506 pKa = 10.25EE507 pKa = 4.64VKK509 pKa = 9.62ADD511 pKa = 4.25YY512 pKa = 10.05YY513 pKa = 11.52NSWQALLYY521 pKa = 10.55DD522 pKa = 4.03VNGNLNGFRR531 pKa = 11.84DD532 pKa = 3.65LTTNKK537 pKa = 9.29TYY539 pKa = 10.09TIRR542 pKa = 11.84SCYY545 pKa = 9.26SGRR548 pKa = 11.84VSAAYY553 pKa = 9.67HH554 pKa = 6.17KK555 pKa = 8.08EE556 pKa = 3.96APEE559 pKa = 3.78PALLYY564 pKa = 10.9RR565 pKa = 11.84NINCSYY571 pKa = 10.95VFTNNISRR579 pKa = 11.84EE580 pKa = 4.24EE581 pKa = 3.99NPLNYY586 pKa = 9.38FDD588 pKa = 5.71SYY590 pKa = 10.97LGCVVNADD598 pKa = 3.38NRR600 pKa = 11.84TDD602 pKa = 3.5EE603 pKa = 5.07ALPNCDD609 pKa = 3.43LRR611 pKa = 11.84MGAGLCVDD619 pKa = 3.67YY620 pKa = 11.11SKK622 pKa = 11.14SRR624 pKa = 11.84RR625 pKa = 11.84ARR627 pKa = 11.84RR628 pKa = 11.84SVSTGYY634 pKa = 11.01RR635 pKa = 11.84LTTFEE640 pKa = 5.37PYY642 pKa = 9.99MPMLVNDD649 pKa = 4.28SVQSVGGLYY658 pKa = 10.45EE659 pKa = 3.97MQIPTNFTIGHH670 pKa = 6.13HH671 pKa = 5.77EE672 pKa = 4.09EE673 pKa = 4.92FIQIRR678 pKa = 11.84APKK681 pKa = 8.75VTIDD685 pKa = 3.67CAAFVCGDD693 pKa = 3.4NAACRR698 pKa = 11.84QQLVEE703 pKa = 4.55YY704 pKa = 10.87GSFCDD709 pKa = 4.0NVNAILNEE717 pKa = 3.97VNNLLDD723 pKa = 4.08NMQLQVASALMQGVTISSRR742 pKa = 11.84LPDD745 pKa = 5.17GISGPIDD752 pKa = 4.86DD753 pKa = 5.34INFSPLLGCIGSTCAEE769 pKa = 4.69DD770 pKa = 4.47GNGPSAIRR778 pKa = 11.84GRR780 pKa = 11.84SAIEE784 pKa = 4.11DD785 pKa = 3.76LLFDD789 pKa = 4.06KK790 pKa = 11.25VKK792 pKa = 10.82LSDD795 pKa = 3.34VGFVEE800 pKa = 6.19AYY802 pKa = 9.87NNCTGGQEE810 pKa = 4.32VRR812 pKa = 11.84DD813 pKa = 3.97LLCVQSFNGIKK824 pKa = 10.05VLPPVLSEE832 pKa = 4.18SQISGYY838 pKa = 7.58TAGATAAAMFPPWTAAAGVPFSLNVQYY865 pKa = 10.59RR866 pKa = 11.84INGLGVTMNVLSEE879 pKa = 4.11NQKK882 pKa = 10.06MIASAFNNALGAIQEE897 pKa = 4.62GFDD900 pKa = 3.73ATNSALGKK908 pKa = 9.37IQSVVNANAEE918 pKa = 4.13ALNNLLNQLSNRR930 pKa = 11.84FGAISASLQEE940 pKa = 4.0ILTRR944 pKa = 11.84LDD946 pKa = 3.15AVEE949 pKa = 5.13AKK951 pKa = 10.57AQIDD955 pKa = 3.56RR956 pKa = 11.84LINGRR961 pKa = 11.84LTALNAYY968 pKa = 9.21ISKK971 pKa = 9.99QLSDD975 pKa = 3.23STLIKK980 pKa = 10.46FSAAQAIEE988 pKa = 4.11KK989 pKa = 10.61VNEE992 pKa = 4.28CVKK995 pKa = 10.93SQTTRR1000 pKa = 11.84INFCGNGNHH1009 pKa = 6.86ILSLVQNAPYY1019 pKa = 9.9GLCFIHH1025 pKa = 6.84FSYY1028 pKa = 11.19VPTSFKK1034 pKa = 9.67TANVSPGLCISGDD1047 pKa = 3.15RR1048 pKa = 11.84GLAPKK1053 pKa = 10.3AGYY1056 pKa = 8.93FVQDD1060 pKa = 3.0NGEE1063 pKa = 4.07WKK1065 pKa = 8.91FTGSNYY1071 pKa = 10.17YY1072 pKa = 9.98YY1073 pKa = 9.83PEE1075 pKa = 5.68PITDD1079 pKa = 3.59KK1080 pKa = 10.85NSVVMISCAVNYY1092 pKa = 7.8TKK1094 pKa = 10.77APEE1097 pKa = 4.02VFLNNSIPNLPDD1109 pKa = 3.66FKK1111 pKa = 11.37EE1112 pKa = 4.4EE1113 pKa = 3.64LDD1115 pKa = 3.46KK1116 pKa = 11.03WFKK1119 pKa = 10.55NQTSIAPDD1127 pKa = 3.69LSLDD1131 pKa = 3.81FEE1133 pKa = 4.58KK1134 pKa = 11.4LNVTFLDD1141 pKa = 3.44LTYY1144 pKa = 11.03EE1145 pKa = 4.04MNRR1148 pKa = 11.84IQDD1151 pKa = 4.14AIKK1154 pKa = 10.34KK1155 pKa = 10.05LNEE1158 pKa = 3.82SYY1160 pKa = 11.14INLKK1164 pKa = 10.23EE1165 pKa = 3.97VGTYY1169 pKa = 9.32EE1170 pKa = 5.21MYY1172 pKa = 11.04VKK1174 pKa = 9.96WPWYY1178 pKa = 8.24VWLLIGLAGVAVCVLLFFICCCTGCGSCCFRR1209 pKa = 11.84KK1210 pKa = 10.28CGSCCDD1216 pKa = 3.87EE1217 pKa = 4.38YY1218 pKa = 11.48GGHH1221 pKa = 6.11QDD1223 pKa = 4.52SIVIHH1228 pKa = 6.54NISAHH1233 pKa = 5.82EE1234 pKa = 4.06DD1235 pKa = 3.09
Molecular weight: 136.68 kDa
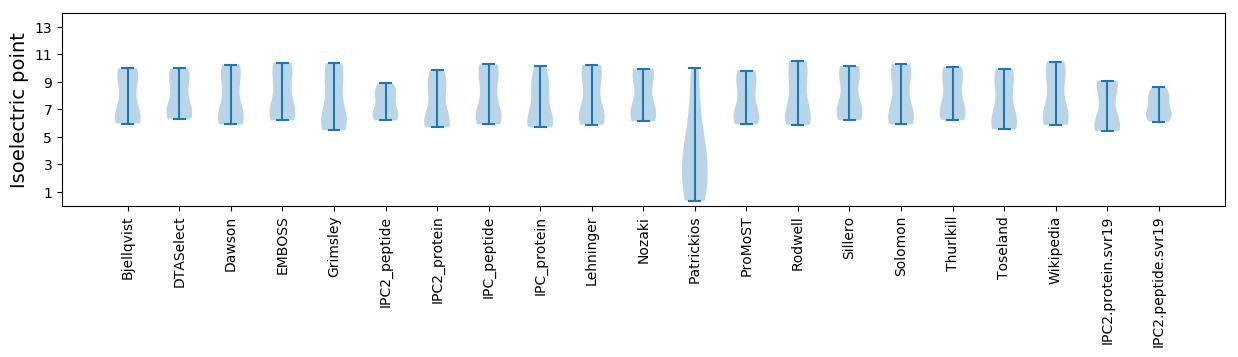
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I7AB47|I7AB47_9BETC Hemagglutinin-esterase OS=Murine coronavirus OX=694005 GN=HE PE=3 SV=1
MM1 pKa = 7.46ALIGPKK7 pKa = 7.57TTIAAVFIGPFLVACMLGIGLVYY30 pKa = 10.81LLQLQVQIFHH40 pKa = 6.45VKK42 pKa = 9.07DD43 pKa = 3.89TIRR46 pKa = 11.84VTGKK50 pKa = 9.2PATVSYY56 pKa = 7.5TTSTPVTPVATTLDD70 pKa = 3.53GTTYY74 pKa = 10.5TLIRR78 pKa = 11.84PTSSYY83 pKa = 9.16TRR85 pKa = 11.84VYY87 pKa = 10.18LGSSRR92 pKa = 11.84GFDD95 pKa = 2.9TSTFGPKK102 pKa = 9.08TLDD105 pKa = 3.78YY106 pKa = 9.39ITSSKK111 pKa = 10.15PHH113 pKa = 6.34LNSGRR118 pKa = 11.84PYY120 pKa = 10.28TLRR123 pKa = 11.84HH124 pKa = 5.44LPKK127 pKa = 10.79YY128 pKa = 6.04MTPPATWRR136 pKa = 11.84FGLL139 pKa = 4.06
MM1 pKa = 7.46ALIGPKK7 pKa = 7.57TTIAAVFIGPFLVACMLGIGLVYY30 pKa = 10.81LLQLQVQIFHH40 pKa = 6.45VKK42 pKa = 9.07DD43 pKa = 3.89TIRR46 pKa = 11.84VTGKK50 pKa = 9.2PATVSYY56 pKa = 7.5TTSTPVTPVATTLDD70 pKa = 3.53GTTYY74 pKa = 10.5TLIRR78 pKa = 11.84PTSSYY83 pKa = 9.16TRR85 pKa = 11.84VYY87 pKa = 10.18LGSSRR92 pKa = 11.84GFDD95 pKa = 2.9TSTFGPKK102 pKa = 9.08TLDD105 pKa = 3.78YY106 pKa = 9.39ITSSKK111 pKa = 10.15PHH113 pKa = 6.34LNSGRR118 pKa = 11.84PYY120 pKa = 10.28TLRR123 pKa = 11.84HH124 pKa = 5.44LPKK127 pKa = 10.79YY128 pKa = 6.04MTPPATWRR136 pKa = 11.84FGLL139 pKa = 4.06
Molecular weight: 15.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14344 |
88 |
7180 |
1593.8 |
177.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.965 ± 0.17 | 3.758 ± 0.249 |
5.556 ± 0.333 | 4.037 ± 0.23 |
5.243 ± 0.109 | 6.177 ± 0.274 |
1.645 ± 0.158 | 4.19 ± 0.415 |
5.779 ± 0.38 | 9.126 ± 0.242 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.363 ± 0.192 | 4.971 ± 0.596 |
3.841 ± 0.293 | 3.437 ± 0.297 |
3.681 ± 0.251 | 7.139 ± 0.15 |
5.752 ± 0.345 | 10.213 ± 0.815 |
1.339 ± 0.108 | 4.769 ± 0.286 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
