
Rhizobium sp. 24NR
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
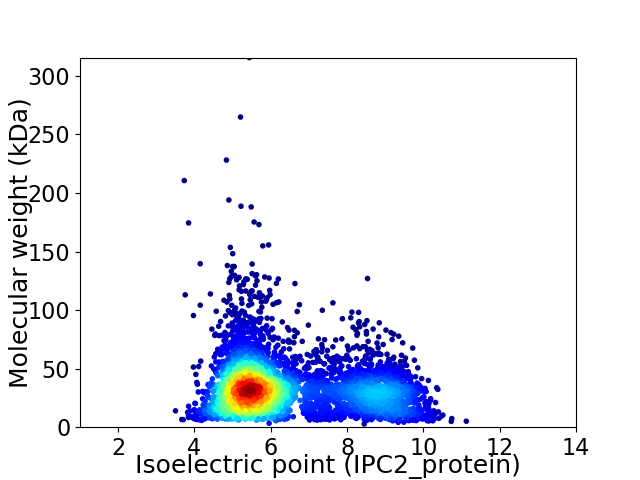
Virtual 2D-PAGE plot for 4834 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A444LLJ9|A0A444LLJ9_9RHIZ ABC transporter permease OS=Rhizobium sp. 24NR OX=2503024 GN=EPK99_02170 PE=3 SV=1
MM1 pKa = 7.77AFQSVRR7 pKa = 11.84KK8 pKa = 8.23VSEE11 pKa = 3.77VARR14 pKa = 11.84YY15 pKa = 7.66VQYY18 pKa = 11.1AIAVAFLYY26 pKa = 10.9LLIPAFGVQAATSPYY41 pKa = 8.54CTNLSANVTQGSSVVIDD58 pKa = 3.61VSNCDD63 pKa = 3.3GPIGLGIGWNGAQPAHH79 pKa = 5.3GTIVIGNQTGPQGMQSVTYY98 pKa = 9.33TPNSNNTATSDD109 pKa = 3.66SFPLEE114 pKa = 4.45DD115 pKa = 3.61EE116 pKa = 4.52NGDD119 pKa = 3.7VFTVNITITPATSASIAVSPSSANEE144 pKa = 3.71DD145 pKa = 3.29SGATFTYY152 pKa = 10.03TVTLSQTSNSATTVNLTRR170 pKa = 11.84SGTATSGTDD179 pKa = 3.11YY180 pKa = 10.54TGAVSSIVVPANTTTASFVVTPVADD205 pKa = 3.61AAVEE209 pKa = 3.8ADD211 pKa = 3.67EE212 pKa = 4.49TVVISVASGSGYY224 pKa = 10.7SIGNPSSATATILNDD239 pKa = 3.88DD240 pKa = 4.56APSASIAVSPSSVAEE255 pKa = 4.38DD256 pKa = 3.62GATNLIYY263 pKa = 10.31TVTLSQAPVTAVSVAFNVGGTATSGSDD290 pKa = 3.46YY291 pKa = 11.15AAVSSPLVIAAGQTSGTITINPTADD316 pKa = 3.24SAVEE320 pKa = 3.7ADD322 pKa = 3.51EE323 pKa = 4.63TVILTLAAGSGYY335 pKa = 10.74SVGSPNSATGTILNDD350 pKa = 3.75DD351 pKa = 4.08QPSLTINDD359 pKa = 3.41VSITEE364 pKa = 4.39GNAGTSTATFTVSLNQPAGAGGVSFDD390 pKa = 3.7IATADD395 pKa = 3.52GTATAGVDD403 pKa = 3.77YY404 pKa = 10.64VASSLSRR411 pKa = 11.84TIAAGSSSATFTVLINGDD429 pKa = 3.84TLNEE433 pKa = 4.03PNEE436 pKa = 4.27TFFVNVSNVTGATVADD452 pKa = 3.91AQGQGTIVNDD462 pKa = 3.96DD463 pKa = 4.0SAPSLAIDD471 pKa = 3.5DD472 pKa = 4.15VSVNEE477 pKa = 4.47GNSGTTTATFTVSLSAASGQTVSVSYY503 pKa = 9.31ATVDD507 pKa = 3.53GTASAGSDD515 pKa = 3.26YY516 pKa = 10.87VARR519 pKa = 11.84SGTLTFAPGITAQGVAVTVNGDD541 pKa = 3.37TSVEE545 pKa = 4.05PNEE548 pKa = 4.28TFSVGLSGASNASIATGTGTGTGTGTGTGTINNDD582 pKa = 3.47DD583 pKa = 3.74AVVTVGPTTLPSATAASAYY602 pKa = 9.99NQTLTASGGTSPYY615 pKa = 10.93AFLVSAGALPSGLTLSSAGVLSGTPTASGSFNFTATATDD654 pKa = 3.43SGGSPTSGSRR664 pKa = 11.84AYY666 pKa = 8.72TLAVAGPTVDD676 pKa = 5.57LPATTLTGGVTGQAYY691 pKa = 9.42AASINPATGGIAPYY705 pKa = 9.83TYY707 pKa = 10.5TLGNGALPAGIVVDD721 pKa = 4.18SASGALSGTPTAAGTFNFTLTATDD745 pKa = 3.69STAGTPSQASRR756 pKa = 11.84SYY758 pKa = 10.99SLTVGQPSLSISPPTLDD775 pKa = 3.3AAVIGSPYY783 pKa = 10.74SKK785 pKa = 10.78VLVGSGGTAPYY796 pKa = 10.78SFVLGSGALPAGLTLAGNGTLSGTPTANGNFSFDD830 pKa = 3.34ATVTDD835 pKa = 4.61AYY837 pKa = 10.65GATASKK843 pKa = 10.8SYY845 pKa = 9.55TLAVNALPSLSIGDD859 pKa = 3.36ISVNEE864 pKa = 4.32GSSGTSIANLTVEE877 pKa = 4.96LSTASGQTVTVNYY890 pKa = 8.07ATADD894 pKa = 3.67GTASAGSDD902 pKa = 3.34YY903 pKa = 11.07VAASGALTFAPGVTTRR919 pKa = 11.84TIAVSINSDD928 pKa = 3.72TNPEE932 pKa = 3.79ASEE935 pKa = 4.32TFSVALSGAVNATIARR951 pKa = 11.84ATGTATIVDD960 pKa = 4.09DD961 pKa = 4.64DD962 pKa = 4.7APVTVGPASLSAATVNAPYY981 pKa = 10.2SQSLTATGGTAPYY994 pKa = 9.23TYY996 pKa = 10.93GVTTGSLPVGLTLSSAGALSGTPTTTGSFSFTASATDD1033 pKa = 3.19SSTPPSTASRR1043 pKa = 11.84SYY1045 pKa = 10.73SIAVSGPVMTLPATTLADD1063 pKa = 3.34AVTGQAYY1070 pKa = 9.95SAAITPATGGTAPYY1084 pKa = 9.12TYY1086 pKa = 10.45AVSAGTLPAGIILNSADD1103 pKa = 3.49GSLSGTPTASGTFTFTLTATDD1124 pKa = 3.82STGGTASQASRR1135 pKa = 11.84SYY1137 pKa = 10.58SLAVGTPSLSVSPPTLDD1154 pKa = 3.17AAVNGRR1160 pKa = 11.84AYY1162 pKa = 10.96SKK1164 pKa = 10.99VLAGSGGTAPYY1175 pKa = 10.12TFLLGSGSLPAGLTLTSNGALSGTPSVNGSFSFNVTVTDD1214 pKa = 3.27THH1216 pKa = 6.64GATGSQSYY1224 pKa = 8.03TLEE1227 pKa = 3.77VKK1229 pKa = 10.79APVGFVFSPSGGALPEE1245 pKa = 3.99AMIGEE1250 pKa = 4.97AYY1252 pKa = 9.85SANISATGGTGALIYY1267 pKa = 10.49SLASGTMPDD1276 pKa = 3.77GLVLNISTGQLNGPLSEE1293 pKa = 4.04AAKK1296 pKa = 10.04SGEE1299 pKa = 3.93YY1300 pKa = 10.27SFSIGVRR1307 pKa = 11.84DD1308 pKa = 3.85SNGNTGSASYY1318 pKa = 9.83VVKK1321 pKa = 10.8VIEE1324 pKa = 4.19RR1325 pKa = 11.84TVTVTNKK1332 pKa = 9.62VVDD1335 pKa = 4.34VPAGTVPPNVYY1346 pKa = 10.26LNRR1349 pKa = 11.84GATGGPFVDD1358 pKa = 3.61ADD1360 pKa = 4.07VIAVEE1365 pKa = 4.24PANAGSIQIIRR1376 pKa = 11.84GEE1378 pKa = 4.19VAQVGPVTPVGFYY1391 pKa = 10.97LKK1393 pKa = 10.3FIPNPAFSGQVNVGYY1408 pKa = 10.56RR1409 pKa = 11.84LISALGVSNQGLVTYY1424 pKa = 10.68NVGLNAAAVAEE1435 pKa = 5.06DD1436 pKa = 4.03IDD1438 pKa = 4.26TLVHH1442 pKa = 6.41GFVQTRR1448 pKa = 11.84QNLIASTINVPGLLEE1463 pKa = 3.84RR1464 pKa = 11.84RR1465 pKa = 11.84RR1466 pKa = 11.84MEE1468 pKa = 4.11EE1469 pKa = 3.37ATDD1472 pKa = 3.8PVTLRR1477 pKa = 11.84MNPDD1481 pKa = 3.43AEE1483 pKa = 4.26GMTASFSTSLVQLQGARR1500 pKa = 11.84DD1501 pKa = 3.57NADD1504 pKa = 3.39GLKK1507 pKa = 10.37SGASSPFNIWLEE1519 pKa = 3.98GTMIFHH1525 pKa = 6.99NRR1527 pKa = 11.84DD1528 pKa = 3.16EE1529 pKa = 5.26NGGKK1533 pKa = 8.83WGNFSMLSTGADD1545 pKa = 3.35YY1546 pKa = 11.34LLSDD1550 pKa = 4.05KK1551 pKa = 11.05ALVGISFHH1559 pKa = 6.81YY1560 pKa = 11.06DD1561 pKa = 2.56HH1562 pKa = 6.61MTDD1565 pKa = 3.29PTEE1568 pKa = 4.15EE1569 pKa = 4.1DD1570 pKa = 3.95AEE1572 pKa = 4.35LTGNGWLAGPYY1583 pKa = 10.44ASFEE1587 pKa = 3.81IGKK1590 pKa = 10.01GVFFDD1595 pKa = 3.57TSLLYY1600 pKa = 10.82GGSSNDD1606 pKa = 2.61IDD1608 pKa = 3.65TAFFDD1613 pKa = 4.13GTFDD1617 pKa = 3.34TSRR1620 pKa = 11.84WLFDD1624 pKa = 3.77SAIKK1628 pKa = 9.8GQWDD1632 pKa = 3.3IDD1634 pKa = 3.78DD1635 pKa = 4.21ATILTPKK1642 pKa = 9.54LRR1644 pKa = 11.84AMYY1647 pKa = 10.27INEE1650 pKa = 4.13KK1651 pKa = 10.05VDD1653 pKa = 4.35DD1654 pKa = 4.16YY1655 pKa = 11.79SVSNSGGDD1663 pKa = 3.33IVDD1666 pKa = 3.85LAGFTEE1672 pKa = 4.12EE1673 pKa = 3.96QLRR1676 pKa = 11.84VSLGAEE1682 pKa = 3.97IKK1684 pKa = 10.42RR1685 pKa = 11.84EE1686 pKa = 3.92YY1687 pKa = 10.53KK1688 pKa = 10.15LQNGASLTPSLGLTAGYY1705 pKa = 10.63AGLDD1709 pKa = 3.33GDD1711 pKa = 4.69GLFGSLAAGISLSTDD1726 pKa = 3.43TAWTFEE1732 pKa = 4.03AGVLFNIEE1740 pKa = 3.84GDD1742 pKa = 4.07GQQSTGAKK1750 pKa = 9.42ISARR1754 pKa = 11.84SQFF1757 pKa = 3.26
MM1 pKa = 7.77AFQSVRR7 pKa = 11.84KK8 pKa = 8.23VSEE11 pKa = 3.77VARR14 pKa = 11.84YY15 pKa = 7.66VQYY18 pKa = 11.1AIAVAFLYY26 pKa = 10.9LLIPAFGVQAATSPYY41 pKa = 8.54CTNLSANVTQGSSVVIDD58 pKa = 3.61VSNCDD63 pKa = 3.3GPIGLGIGWNGAQPAHH79 pKa = 5.3GTIVIGNQTGPQGMQSVTYY98 pKa = 9.33TPNSNNTATSDD109 pKa = 3.66SFPLEE114 pKa = 4.45DD115 pKa = 3.61EE116 pKa = 4.52NGDD119 pKa = 3.7VFTVNITITPATSASIAVSPSSANEE144 pKa = 3.71DD145 pKa = 3.29SGATFTYY152 pKa = 10.03TVTLSQTSNSATTVNLTRR170 pKa = 11.84SGTATSGTDD179 pKa = 3.11YY180 pKa = 10.54TGAVSSIVVPANTTTASFVVTPVADD205 pKa = 3.61AAVEE209 pKa = 3.8ADD211 pKa = 3.67EE212 pKa = 4.49TVVISVASGSGYY224 pKa = 10.7SIGNPSSATATILNDD239 pKa = 3.88DD240 pKa = 4.56APSASIAVSPSSVAEE255 pKa = 4.38DD256 pKa = 3.62GATNLIYY263 pKa = 10.31TVTLSQAPVTAVSVAFNVGGTATSGSDD290 pKa = 3.46YY291 pKa = 11.15AAVSSPLVIAAGQTSGTITINPTADD316 pKa = 3.24SAVEE320 pKa = 3.7ADD322 pKa = 3.51EE323 pKa = 4.63TVILTLAAGSGYY335 pKa = 10.74SVGSPNSATGTILNDD350 pKa = 3.75DD351 pKa = 4.08QPSLTINDD359 pKa = 3.41VSITEE364 pKa = 4.39GNAGTSTATFTVSLNQPAGAGGVSFDD390 pKa = 3.7IATADD395 pKa = 3.52GTATAGVDD403 pKa = 3.77YY404 pKa = 10.64VASSLSRR411 pKa = 11.84TIAAGSSSATFTVLINGDD429 pKa = 3.84TLNEE433 pKa = 4.03PNEE436 pKa = 4.27TFFVNVSNVTGATVADD452 pKa = 3.91AQGQGTIVNDD462 pKa = 3.96DD463 pKa = 4.0SAPSLAIDD471 pKa = 3.5DD472 pKa = 4.15VSVNEE477 pKa = 4.47GNSGTTTATFTVSLSAASGQTVSVSYY503 pKa = 9.31ATVDD507 pKa = 3.53GTASAGSDD515 pKa = 3.26YY516 pKa = 10.87VARR519 pKa = 11.84SGTLTFAPGITAQGVAVTVNGDD541 pKa = 3.37TSVEE545 pKa = 4.05PNEE548 pKa = 4.28TFSVGLSGASNASIATGTGTGTGTGTGTGTINNDD582 pKa = 3.47DD583 pKa = 3.74AVVTVGPTTLPSATAASAYY602 pKa = 9.99NQTLTASGGTSPYY615 pKa = 10.93AFLVSAGALPSGLTLSSAGVLSGTPTASGSFNFTATATDD654 pKa = 3.43SGGSPTSGSRR664 pKa = 11.84AYY666 pKa = 8.72TLAVAGPTVDD676 pKa = 5.57LPATTLTGGVTGQAYY691 pKa = 9.42AASINPATGGIAPYY705 pKa = 9.83TYY707 pKa = 10.5TLGNGALPAGIVVDD721 pKa = 4.18SASGALSGTPTAAGTFNFTLTATDD745 pKa = 3.69STAGTPSQASRR756 pKa = 11.84SYY758 pKa = 10.99SLTVGQPSLSISPPTLDD775 pKa = 3.3AAVIGSPYY783 pKa = 10.74SKK785 pKa = 10.78VLVGSGGTAPYY796 pKa = 10.78SFVLGSGALPAGLTLAGNGTLSGTPTANGNFSFDD830 pKa = 3.34ATVTDD835 pKa = 4.61AYY837 pKa = 10.65GATASKK843 pKa = 10.8SYY845 pKa = 9.55TLAVNALPSLSIGDD859 pKa = 3.36ISVNEE864 pKa = 4.32GSSGTSIANLTVEE877 pKa = 4.96LSTASGQTVTVNYY890 pKa = 8.07ATADD894 pKa = 3.67GTASAGSDD902 pKa = 3.34YY903 pKa = 11.07VAASGALTFAPGVTTRR919 pKa = 11.84TIAVSINSDD928 pKa = 3.72TNPEE932 pKa = 3.79ASEE935 pKa = 4.32TFSVALSGAVNATIARR951 pKa = 11.84ATGTATIVDD960 pKa = 4.09DD961 pKa = 4.64DD962 pKa = 4.7APVTVGPASLSAATVNAPYY981 pKa = 10.2SQSLTATGGTAPYY994 pKa = 9.23TYY996 pKa = 10.93GVTTGSLPVGLTLSSAGALSGTPTTTGSFSFTASATDD1033 pKa = 3.19SSTPPSTASRR1043 pKa = 11.84SYY1045 pKa = 10.73SIAVSGPVMTLPATTLADD1063 pKa = 3.34AVTGQAYY1070 pKa = 9.95SAAITPATGGTAPYY1084 pKa = 9.12TYY1086 pKa = 10.45AVSAGTLPAGIILNSADD1103 pKa = 3.49GSLSGTPTASGTFTFTLTATDD1124 pKa = 3.82STGGTASQASRR1135 pKa = 11.84SYY1137 pKa = 10.58SLAVGTPSLSVSPPTLDD1154 pKa = 3.17AAVNGRR1160 pKa = 11.84AYY1162 pKa = 10.96SKK1164 pKa = 10.99VLAGSGGTAPYY1175 pKa = 10.12TFLLGSGSLPAGLTLTSNGALSGTPSVNGSFSFNVTVTDD1214 pKa = 3.27THH1216 pKa = 6.64GATGSQSYY1224 pKa = 8.03TLEE1227 pKa = 3.77VKK1229 pKa = 10.79APVGFVFSPSGGALPEE1245 pKa = 3.99AMIGEE1250 pKa = 4.97AYY1252 pKa = 9.85SANISATGGTGALIYY1267 pKa = 10.49SLASGTMPDD1276 pKa = 3.77GLVLNISTGQLNGPLSEE1293 pKa = 4.04AAKK1296 pKa = 10.04SGEE1299 pKa = 3.93YY1300 pKa = 10.27SFSIGVRR1307 pKa = 11.84DD1308 pKa = 3.85SNGNTGSASYY1318 pKa = 9.83VVKK1321 pKa = 10.8VIEE1324 pKa = 4.19RR1325 pKa = 11.84TVTVTNKK1332 pKa = 9.62VVDD1335 pKa = 4.34VPAGTVPPNVYY1346 pKa = 10.26LNRR1349 pKa = 11.84GATGGPFVDD1358 pKa = 3.61ADD1360 pKa = 4.07VIAVEE1365 pKa = 4.24PANAGSIQIIRR1376 pKa = 11.84GEE1378 pKa = 4.19VAQVGPVTPVGFYY1391 pKa = 10.97LKK1393 pKa = 10.3FIPNPAFSGQVNVGYY1408 pKa = 10.56RR1409 pKa = 11.84LISALGVSNQGLVTYY1424 pKa = 10.68NVGLNAAAVAEE1435 pKa = 5.06DD1436 pKa = 4.03IDD1438 pKa = 4.26TLVHH1442 pKa = 6.41GFVQTRR1448 pKa = 11.84QNLIASTINVPGLLEE1463 pKa = 3.84RR1464 pKa = 11.84RR1465 pKa = 11.84RR1466 pKa = 11.84MEE1468 pKa = 4.11EE1469 pKa = 3.37ATDD1472 pKa = 3.8PVTLRR1477 pKa = 11.84MNPDD1481 pKa = 3.43AEE1483 pKa = 4.26GMTASFSTSLVQLQGARR1500 pKa = 11.84DD1501 pKa = 3.57NADD1504 pKa = 3.39GLKK1507 pKa = 10.37SGASSPFNIWLEE1519 pKa = 3.98GTMIFHH1525 pKa = 6.99NRR1527 pKa = 11.84DD1528 pKa = 3.16EE1529 pKa = 5.26NGGKK1533 pKa = 8.83WGNFSMLSTGADD1545 pKa = 3.35YY1546 pKa = 11.34LLSDD1550 pKa = 4.05KK1551 pKa = 11.05ALVGISFHH1559 pKa = 6.81YY1560 pKa = 11.06DD1561 pKa = 2.56HH1562 pKa = 6.61MTDD1565 pKa = 3.29PTEE1568 pKa = 4.15EE1569 pKa = 4.1DD1570 pKa = 3.95AEE1572 pKa = 4.35LTGNGWLAGPYY1583 pKa = 10.44ASFEE1587 pKa = 3.81IGKK1590 pKa = 10.01GVFFDD1595 pKa = 3.57TSLLYY1600 pKa = 10.82GGSSNDD1606 pKa = 2.61IDD1608 pKa = 3.65TAFFDD1613 pKa = 4.13GTFDD1617 pKa = 3.34TSRR1620 pKa = 11.84WLFDD1624 pKa = 3.77SAIKK1628 pKa = 9.8GQWDD1632 pKa = 3.3IDD1634 pKa = 3.78DD1635 pKa = 4.21ATILTPKK1642 pKa = 9.54LRR1644 pKa = 11.84AMYY1647 pKa = 10.27INEE1650 pKa = 4.13KK1651 pKa = 10.05VDD1653 pKa = 4.35DD1654 pKa = 4.16YY1655 pKa = 11.79SVSNSGGDD1663 pKa = 3.33IVDD1666 pKa = 3.85LAGFTEE1672 pKa = 4.12EE1673 pKa = 3.96QLRR1676 pKa = 11.84VSLGAEE1682 pKa = 3.97IKK1684 pKa = 10.42RR1685 pKa = 11.84EE1686 pKa = 3.92YY1687 pKa = 10.53KK1688 pKa = 10.15LQNGASLTPSLGLTAGYY1705 pKa = 10.63AGLDD1709 pKa = 3.33GDD1711 pKa = 4.69GLFGSLAAGISLSTDD1726 pKa = 3.43TAWTFEE1732 pKa = 4.03AGVLFNIEE1740 pKa = 3.84GDD1742 pKa = 4.07GQQSTGAKK1750 pKa = 9.42ISARR1754 pKa = 11.84SQFF1757 pKa = 3.26
Molecular weight: 174.47 kDa
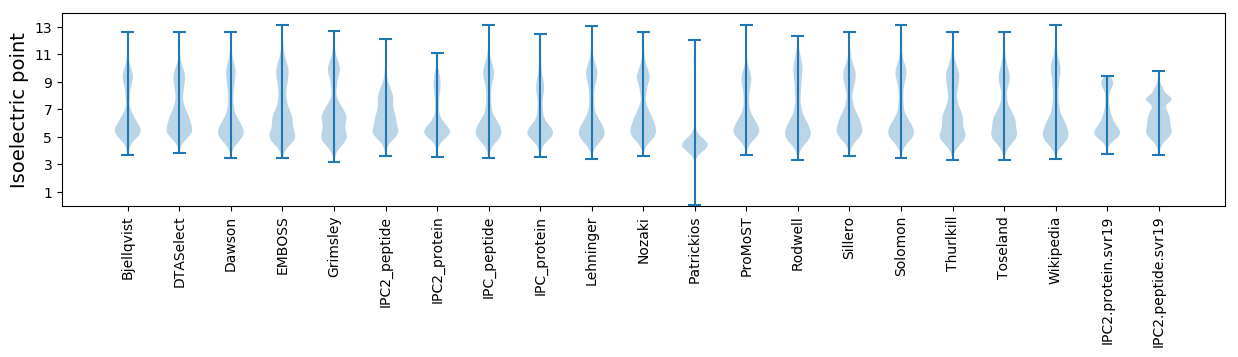
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S3RXQ3|A0A3S3RXQ3_9RHIZ FAD-binding oxidoreductase OS=Rhizobium sp. 24NR OX=2503024 GN=EPK99_01080 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27GGQKK29 pKa = 9.83VLSARR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84GRR39 pKa = 11.84QRR41 pKa = 11.84LSAA44 pKa = 4.11
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27GGQKK29 pKa = 9.83VLSARR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84GRR39 pKa = 11.84QRR41 pKa = 11.84LSAA44 pKa = 4.11
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1524728 |
26 |
2822 |
315.4 |
34.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.707 ± 0.043 | 0.746 ± 0.01 |
5.723 ± 0.027 | 5.727 ± 0.036 |
3.866 ± 0.022 | 8.318 ± 0.03 |
1.992 ± 0.018 | 5.71 ± 0.026 |
3.699 ± 0.025 | 10.056 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.598 ± 0.017 | 2.817 ± 0.017 |
4.93 ± 0.026 | 3.268 ± 0.02 |
6.599 ± 0.034 | 5.99 ± 0.03 |
5.339 ± 0.026 | 7.388 ± 0.026 |
1.247 ± 0.014 | 2.279 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
