
Helicobacter canis NCTC 12740
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Helicobacteraceae; Helicobacter; Helicobacter canis
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
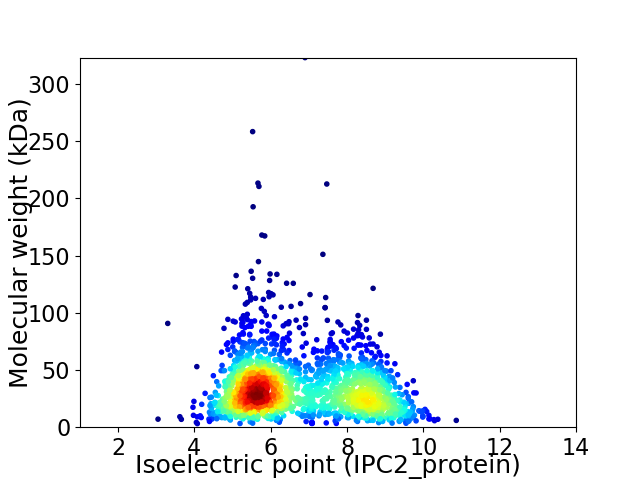
Virtual 2D-PAGE plot for 1800 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
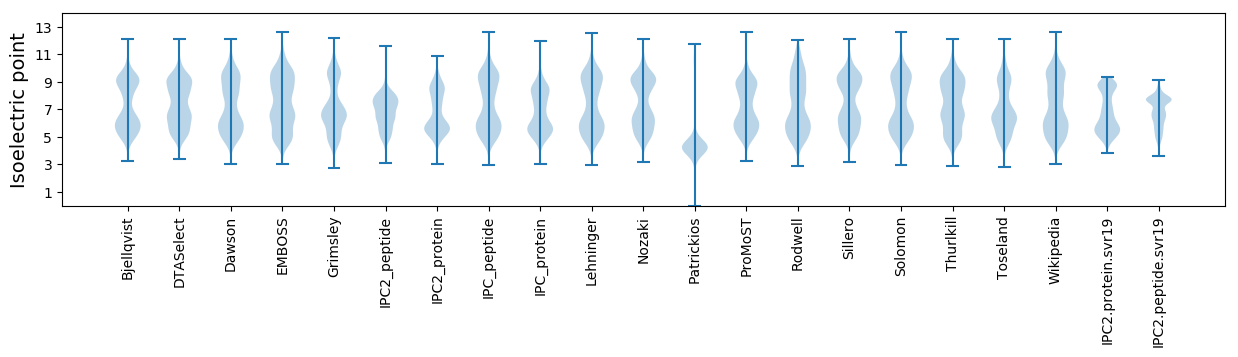
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V8CES6|V8CES6_9HELI Uncharacterized protein OS=Helicobacter canis NCTC 12740 OX=1357399 GN=HMPREF2087_01415 PE=4 SV=1
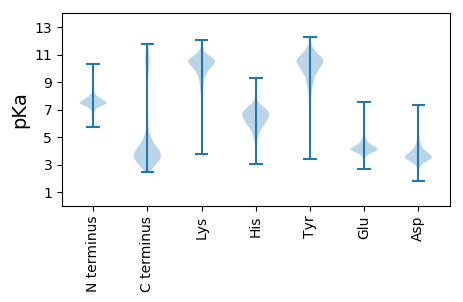
MM1 pKa = 7.52AVLHH5 pKa = 6.94DD6 pKa = 3.87VLNNSDD12 pKa = 4.41YY13 pKa = 11.05MLDD16 pKa = 3.6LCADD20 pKa = 3.75KK21 pKa = 10.96SLTMADD27 pKa = 4.25LDD29 pKa = 3.95SSSVFICRR37 pKa = 11.84CC38 pKa = 3.23
MM1 pKa = 7.52AVLHH5 pKa = 6.94DD6 pKa = 3.87VLNNSDD12 pKa = 4.41YY13 pKa = 11.05MLDD16 pKa = 3.6LCADD20 pKa = 3.75KK21 pKa = 10.96SLTMADD27 pKa = 4.25LDD29 pKa = 3.95SSSVFICRR37 pKa = 11.84CC38 pKa = 3.23
Molecular weight: 4.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V8CIC0|V8CIC0_9HELI Single-stranded-DNA-specific exonuclease RecJ OS=Helicobacter canis NCTC 12740 OX=1357399 GN=HMPREF2087_00061 PE=3 SV=1
MM1 pKa = 8.13DD2 pKa = 4.43SRR4 pKa = 11.84VLTMDD9 pKa = 3.2YY10 pKa = 8.63HH11 pKa = 6.11TLQSKK16 pKa = 9.85ARR18 pKa = 11.84NDD20 pKa = 3.4DD21 pKa = 3.65TNADD25 pKa = 3.31SSMRR29 pKa = 11.84WILAPSFPAHH39 pKa = 6.95APRR42 pKa = 11.84ISPHH46 pKa = 5.04YY47 pKa = 8.8LKK49 pKa = 10.82HH50 pKa = 5.87SATLRR55 pKa = 11.84YY56 pKa = 8.88QALLALQRR64 pKa = 11.84TKK66 pKa = 10.48PCHH69 pKa = 5.1HH70 pKa = 6.5TQANTAHH77 pKa = 7.35KK78 pKa = 9.18PQNLPPKK85 pKa = 8.49HH86 pKa = 5.92RR87 pKa = 11.84HH88 pKa = 5.86IITLGIGANLGTRR101 pKa = 11.84QEE103 pKa = 3.71ICKK106 pKa = 10.23RR107 pKa = 11.84FAHH110 pKa = 6.53LLDD113 pKa = 4.14SLTSKK118 pKa = 11.01DD119 pKa = 3.57FTLLATSPILHH130 pKa = 6.53NPPFGYY136 pKa = 9.39TNQPYY141 pKa = 9.81FYY143 pKa = 10.14NATITLATSLSVVEE157 pKa = 3.94LFARR161 pKa = 11.84VFYY164 pKa = 10.57LEE166 pKa = 3.98RR167 pKa = 11.84RR168 pKa = 11.84FGRR171 pKa = 11.84GRR173 pKa = 11.84KK174 pKa = 8.84RR175 pKa = 11.84AFKK178 pKa = 10.15NAPRR182 pKa = 11.84TLDD185 pKa = 3.06IDD187 pKa = 3.61IIFYY191 pKa = 10.95DD192 pKa = 3.44KK193 pKa = 11.21VIIKK197 pKa = 10.1RR198 pKa = 11.84PYY200 pKa = 8.8LTIPHH205 pKa = 6.57HH206 pKa = 6.03GFRR209 pKa = 11.84TRR211 pKa = 11.84DD212 pKa = 3.49SVLLPLGFMLKK223 pKa = 9.89QVLEE227 pKa = 4.53SKK229 pKa = 10.53SIHH232 pKa = 5.65QSTRR236 pKa = 11.84SKK238 pKa = 10.28PCRR241 pKa = 11.84PLLSQEE247 pKa = 4.33KK248 pKa = 9.67LQQKK252 pKa = 8.89HH253 pKa = 4.35SKK255 pKa = 9.07RR256 pKa = 11.84RR257 pKa = 11.84AMSS260 pKa = 3.28
MM1 pKa = 8.13DD2 pKa = 4.43SRR4 pKa = 11.84VLTMDD9 pKa = 3.2YY10 pKa = 8.63HH11 pKa = 6.11TLQSKK16 pKa = 9.85ARR18 pKa = 11.84NDD20 pKa = 3.4DD21 pKa = 3.65TNADD25 pKa = 3.31SSMRR29 pKa = 11.84WILAPSFPAHH39 pKa = 6.95APRR42 pKa = 11.84ISPHH46 pKa = 5.04YY47 pKa = 8.8LKK49 pKa = 10.82HH50 pKa = 5.87SATLRR55 pKa = 11.84YY56 pKa = 8.88QALLALQRR64 pKa = 11.84TKK66 pKa = 10.48PCHH69 pKa = 5.1HH70 pKa = 6.5TQANTAHH77 pKa = 7.35KK78 pKa = 9.18PQNLPPKK85 pKa = 8.49HH86 pKa = 5.92RR87 pKa = 11.84HH88 pKa = 5.86IITLGIGANLGTRR101 pKa = 11.84QEE103 pKa = 3.71ICKK106 pKa = 10.23RR107 pKa = 11.84FAHH110 pKa = 6.53LLDD113 pKa = 4.14SLTSKK118 pKa = 11.01DD119 pKa = 3.57FTLLATSPILHH130 pKa = 6.53NPPFGYY136 pKa = 9.39TNQPYY141 pKa = 9.81FYY143 pKa = 10.14NATITLATSLSVVEE157 pKa = 3.94LFARR161 pKa = 11.84VFYY164 pKa = 10.57LEE166 pKa = 3.98RR167 pKa = 11.84RR168 pKa = 11.84FGRR171 pKa = 11.84GRR173 pKa = 11.84KK174 pKa = 8.84RR175 pKa = 11.84AFKK178 pKa = 10.15NAPRR182 pKa = 11.84TLDD185 pKa = 3.06IDD187 pKa = 3.61IIFYY191 pKa = 10.95DD192 pKa = 3.44KK193 pKa = 11.21VIIKK197 pKa = 10.1RR198 pKa = 11.84PYY200 pKa = 8.8LTIPHH205 pKa = 6.57HH206 pKa = 6.03GFRR209 pKa = 11.84TRR211 pKa = 11.84DD212 pKa = 3.49SVLLPLGFMLKK223 pKa = 9.89QVLEE227 pKa = 4.53SKK229 pKa = 10.53SIHH232 pKa = 5.65QSTRR236 pKa = 11.84SKK238 pKa = 10.28PCRR241 pKa = 11.84PLLSQEE247 pKa = 4.33KK248 pKa = 9.67LQQKK252 pKa = 8.89HH253 pKa = 4.35SKK255 pKa = 9.07RR256 pKa = 11.84RR257 pKa = 11.84AMSS260 pKa = 3.28
Molecular weight: 29.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
596658 |
29 |
2885 |
331.5 |
36.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.754 ± 0.056 | 1.381 ± 0.025 |
5.092 ± 0.037 | 5.991 ± 0.061 |
4.744 ± 0.046 | 6.191 ± 0.056 |
2.146 ± 0.028 | 7.438 ± 0.05 |
6.641 ± 0.063 | 10.781 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.111 ± 0.027 | 4.047 ± 0.041 |
3.645 ± 0.037 | 4.247 ± 0.039 |
4.293 ± 0.035 | 7.845 ± 0.055 |
4.654 ± 0.036 | 5.537 ± 0.052 |
0.789 ± 0.017 | 3.67 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
