
Sierra dome spider associated circular virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus siedo1
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
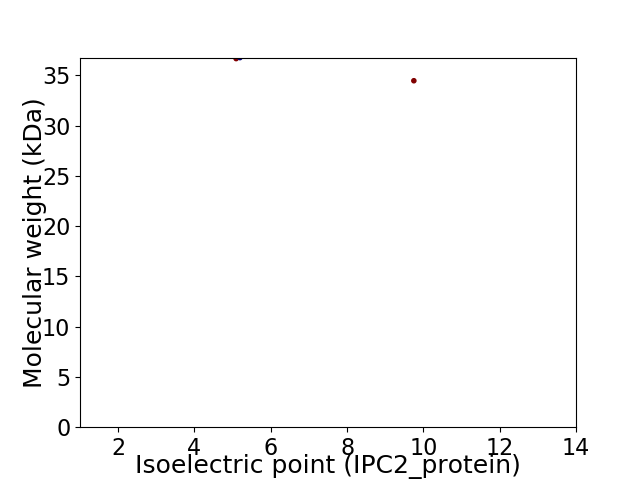
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BP85|A0A346BP85_9VIRU Putative capsid protein OS=Sierra dome spider associated circular virus 1 OX=2293299 PE=4 SV=1
MM1 pKa = 7.4LFVNSKK7 pKa = 10.5YY8 pKa = 11.07VLLTFAQCGDD18 pKa = 3.5LDD20 pKa = 3.67EE21 pKa = 4.61WHH23 pKa = 6.87VSDD26 pKa = 5.14HH27 pKa = 6.37LSSLGAEE34 pKa = 4.27CIVARR39 pKa = 11.84EE40 pKa = 3.99VHH42 pKa = 6.0PTTGGVHH49 pKa = 5.86LHH51 pKa = 5.79VFVDD55 pKa = 4.54FGRR58 pKa = 11.84KK59 pKa = 7.99FRR61 pKa = 11.84SRR63 pKa = 11.84NVRR66 pKa = 11.84VFDD69 pKa = 3.45VDD71 pKa = 3.28GRR73 pKa = 11.84HH74 pKa = 6.11PNVVPSRR81 pKa = 11.84GTPEE85 pKa = 3.88KK86 pKa = 10.92GFDD89 pKa = 3.46YY90 pKa = 10.65AIKK93 pKa = 10.86DD94 pKa = 3.51GDD96 pKa = 4.01VVAGGLEE103 pKa = 3.89RR104 pKa = 11.84PMPRR108 pKa = 11.84GGMSVGAEE116 pKa = 4.17NIRR119 pKa = 11.84NVAGLCEE126 pKa = 4.32STTEE130 pKa = 3.87FLEE133 pKa = 5.37LFDD136 pKa = 4.33EE137 pKa = 4.48MDD139 pKa = 3.58RR140 pKa = 11.84GTLFKK145 pKa = 10.89SFTNIRR151 pKa = 11.84SYY153 pKa = 11.64ADD155 pKa = 2.71WRR157 pKa = 11.84FAVEE161 pKa = 4.11PEE163 pKa = 4.38PYY165 pKa = 9.98VPPFGTEE172 pKa = 3.72FSGGEE177 pKa = 3.68FDD179 pKa = 6.42GRR181 pKa = 11.84DD182 pKa = 3.16DD183 pKa = 3.53WLAQSGIGSKK193 pKa = 10.31SLILYY198 pKa = 8.68GPSRR202 pKa = 11.84TGKK205 pKa = 8.43TSWARR210 pKa = 11.84SLGAHH215 pKa = 7.18LYY217 pKa = 10.21FEE219 pKa = 4.91RR220 pKa = 11.84LFSGKK225 pKa = 8.65EE226 pKa = 4.07AINHH230 pKa = 4.95MSEE233 pKa = 3.66VEE235 pKa = 4.06YY236 pKa = 10.91AVFDD240 pKa = 4.18DD241 pKa = 4.85CSINHH246 pKa = 5.1MPGWKK251 pKa = 9.23SWFGAQATVGVRR263 pKa = 11.84ALYY266 pKa = 10.45RR267 pKa = 11.84DD268 pKa = 3.52ATYY271 pKa = 10.87VAWGKK276 pKa = 9.77PIVWCTNRR284 pKa = 11.84DD285 pKa = 3.12PRR287 pKa = 11.84IDD289 pKa = 3.51MRR291 pKa = 11.84IDD293 pKa = 2.99IDD295 pKa = 3.82NEE297 pKa = 3.89RR298 pKa = 11.84SHH300 pKa = 6.57MWFQDD305 pKa = 3.82DD306 pKa = 4.3IDD308 pKa = 3.78WMEE311 pKa = 4.26ANCVFINITDD321 pKa = 3.57NLYY324 pKa = 10.98
MM1 pKa = 7.4LFVNSKK7 pKa = 10.5YY8 pKa = 11.07VLLTFAQCGDD18 pKa = 3.5LDD20 pKa = 3.67EE21 pKa = 4.61WHH23 pKa = 6.87VSDD26 pKa = 5.14HH27 pKa = 6.37LSSLGAEE34 pKa = 4.27CIVARR39 pKa = 11.84EE40 pKa = 3.99VHH42 pKa = 6.0PTTGGVHH49 pKa = 5.86LHH51 pKa = 5.79VFVDD55 pKa = 4.54FGRR58 pKa = 11.84KK59 pKa = 7.99FRR61 pKa = 11.84SRR63 pKa = 11.84NVRR66 pKa = 11.84VFDD69 pKa = 3.45VDD71 pKa = 3.28GRR73 pKa = 11.84HH74 pKa = 6.11PNVVPSRR81 pKa = 11.84GTPEE85 pKa = 3.88KK86 pKa = 10.92GFDD89 pKa = 3.46YY90 pKa = 10.65AIKK93 pKa = 10.86DD94 pKa = 3.51GDD96 pKa = 4.01VVAGGLEE103 pKa = 3.89RR104 pKa = 11.84PMPRR108 pKa = 11.84GGMSVGAEE116 pKa = 4.17NIRR119 pKa = 11.84NVAGLCEE126 pKa = 4.32STTEE130 pKa = 3.87FLEE133 pKa = 5.37LFDD136 pKa = 4.33EE137 pKa = 4.48MDD139 pKa = 3.58RR140 pKa = 11.84GTLFKK145 pKa = 10.89SFTNIRR151 pKa = 11.84SYY153 pKa = 11.64ADD155 pKa = 2.71WRR157 pKa = 11.84FAVEE161 pKa = 4.11PEE163 pKa = 4.38PYY165 pKa = 9.98VPPFGTEE172 pKa = 3.72FSGGEE177 pKa = 3.68FDD179 pKa = 6.42GRR181 pKa = 11.84DD182 pKa = 3.16DD183 pKa = 3.53WLAQSGIGSKK193 pKa = 10.31SLILYY198 pKa = 8.68GPSRR202 pKa = 11.84TGKK205 pKa = 8.43TSWARR210 pKa = 11.84SLGAHH215 pKa = 7.18LYY217 pKa = 10.21FEE219 pKa = 4.91RR220 pKa = 11.84LFSGKK225 pKa = 8.65EE226 pKa = 4.07AINHH230 pKa = 4.95MSEE233 pKa = 3.66VEE235 pKa = 4.06YY236 pKa = 10.91AVFDD240 pKa = 4.18DD241 pKa = 4.85CSINHH246 pKa = 5.1MPGWKK251 pKa = 9.23SWFGAQATVGVRR263 pKa = 11.84ALYY266 pKa = 10.45RR267 pKa = 11.84DD268 pKa = 3.52ATYY271 pKa = 10.87VAWGKK276 pKa = 9.77PIVWCTNRR284 pKa = 11.84DD285 pKa = 3.12PRR287 pKa = 11.84IDD289 pKa = 3.51MRR291 pKa = 11.84IDD293 pKa = 2.99IDD295 pKa = 3.82NEE297 pKa = 3.89RR298 pKa = 11.84SHH300 pKa = 6.57MWFQDD305 pKa = 3.82DD306 pKa = 4.3IDD308 pKa = 3.78WMEE311 pKa = 4.26ANCVFINITDD321 pKa = 3.57NLYY324 pKa = 10.98
Molecular weight: 36.68 kDa
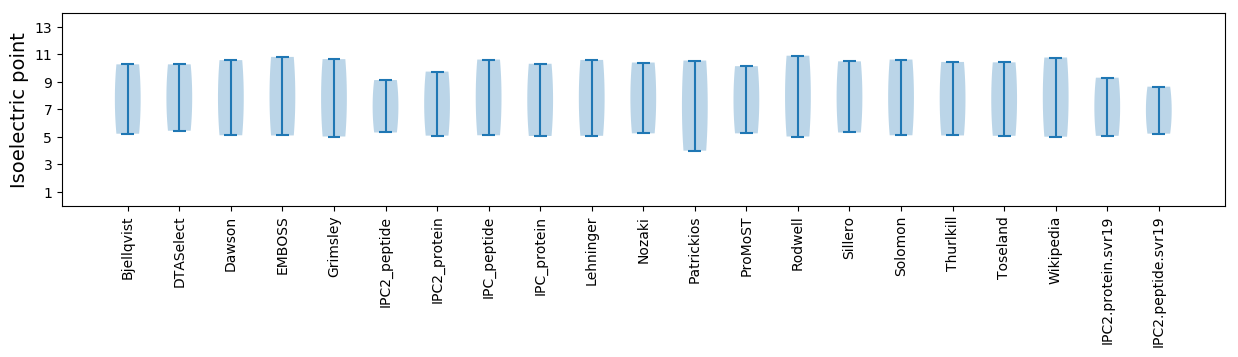
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BP85|A0A346BP85_9VIRU Putative capsid protein OS=Sierra dome spider associated circular virus 1 OX=2293299 PE=4 SV=1
MM1 pKa = 7.36RR2 pKa = 11.84TRR4 pKa = 11.84ARR6 pKa = 11.84RR7 pKa = 11.84TRR9 pKa = 11.84RR10 pKa = 11.84SFARR14 pKa = 11.84SKK16 pKa = 10.46RR17 pKa = 11.84PRR19 pKa = 11.84RR20 pKa = 11.84SYY22 pKa = 10.44LPSRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84SGGKK32 pKa = 9.24KK33 pKa = 7.49RR34 pKa = 11.84TYY36 pKa = 9.8RR37 pKa = 11.84KK38 pKa = 9.55KK39 pKa = 10.64GSRR42 pKa = 11.84KK43 pKa = 8.87MILNMTSTKK52 pKa = 10.4KK53 pKa = 10.01RR54 pKa = 11.84DD55 pKa = 3.25TMINYY60 pKa = 7.93TNVTGTSQIGGTAYY74 pKa = 9.13STSPAIITGGYY85 pKa = 4.77TTFNPFVWVATARR98 pKa = 11.84DD99 pKa = 3.67NTTANSGGAGLKK111 pKa = 9.81FFQSTRR117 pKa = 11.84TATTCFMRR125 pKa = 11.84GLAEE129 pKa = 4.35SIEE132 pKa = 4.19IQVSDD137 pKa = 3.79GLPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTYY150 pKa = 10.31KK151 pKa = 10.72GIADD155 pKa = 5.96LIPSVTTAPAFSLSQEE171 pKa = 4.33TSSGWVRR178 pKa = 11.84TVNALPNNTYY188 pKa = 10.28RR189 pKa = 11.84DD190 pKa = 3.51NFEE193 pKa = 4.11AVIFRR198 pKa = 11.84GSKK201 pKa = 10.36GVDD204 pKa = 3.72FSEE207 pKa = 4.89PLTAPLDD214 pKa = 3.7PTRR217 pKa = 11.84ITVKK221 pKa = 10.16YY222 pKa = 10.36DD223 pKa = 3.0KK224 pKa = 10.49TRR226 pKa = 11.84TIASGNEE233 pKa = 3.71DD234 pKa = 3.36GCIRR238 pKa = 11.84KK239 pKa = 9.63YY240 pKa = 10.98KK241 pKa = 9.76FWHH244 pKa = 5.91GMNKK248 pKa = 9.61NLVYY252 pKa = 10.79ADD254 pKa = 4.34DD255 pKa = 4.01EE256 pKa = 5.04SGGVEE261 pKa = 3.72NVGYY265 pKa = 10.59FSTQSKK271 pKa = 10.98AGMGDD276 pKa = 3.82YY277 pKa = 10.98VVVDD281 pKa = 3.39YY282 pKa = 8.86FLPRR286 pKa = 11.84TGSTSANKK294 pKa = 10.38LSFNATASLYY304 pKa = 8.39WHH306 pKa = 7.02EE307 pKa = 4.37KK308 pKa = 9.19
MM1 pKa = 7.36RR2 pKa = 11.84TRR4 pKa = 11.84ARR6 pKa = 11.84RR7 pKa = 11.84TRR9 pKa = 11.84RR10 pKa = 11.84SFARR14 pKa = 11.84SKK16 pKa = 10.46RR17 pKa = 11.84PRR19 pKa = 11.84RR20 pKa = 11.84SYY22 pKa = 10.44LPSRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84SGGKK32 pKa = 9.24KK33 pKa = 7.49RR34 pKa = 11.84TYY36 pKa = 9.8RR37 pKa = 11.84KK38 pKa = 9.55KK39 pKa = 10.64GSRR42 pKa = 11.84KK43 pKa = 8.87MILNMTSTKK52 pKa = 10.4KK53 pKa = 10.01RR54 pKa = 11.84DD55 pKa = 3.25TMINYY60 pKa = 7.93TNVTGTSQIGGTAYY74 pKa = 9.13STSPAIITGGYY85 pKa = 4.77TTFNPFVWVATARR98 pKa = 11.84DD99 pKa = 3.67NTTANSGGAGLKK111 pKa = 9.81FFQSTRR117 pKa = 11.84TATTCFMRR125 pKa = 11.84GLAEE129 pKa = 4.35SIEE132 pKa = 4.19IQVSDD137 pKa = 3.79GLPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTYY150 pKa = 10.31KK151 pKa = 10.72GIADD155 pKa = 5.96LIPSVTTAPAFSLSQEE171 pKa = 4.33TSSGWVRR178 pKa = 11.84TVNALPNNTYY188 pKa = 10.28RR189 pKa = 11.84DD190 pKa = 3.51NFEE193 pKa = 4.11AVIFRR198 pKa = 11.84GSKK201 pKa = 10.36GVDD204 pKa = 3.72FSEE207 pKa = 4.89PLTAPLDD214 pKa = 3.7PTRR217 pKa = 11.84ITVKK221 pKa = 10.16YY222 pKa = 10.36DD223 pKa = 3.0KK224 pKa = 10.49TRR226 pKa = 11.84TIASGNEE233 pKa = 3.71DD234 pKa = 3.36GCIRR238 pKa = 11.84KK239 pKa = 9.63YY240 pKa = 10.98KK241 pKa = 9.76FWHH244 pKa = 5.91GMNKK248 pKa = 9.61NLVYY252 pKa = 10.79ADD254 pKa = 4.34DD255 pKa = 4.01EE256 pKa = 5.04SGGVEE261 pKa = 3.72NVGYY265 pKa = 10.59FSTQSKK271 pKa = 10.98AGMGDD276 pKa = 3.82YY277 pKa = 10.98VVVDD281 pKa = 3.39YY282 pKa = 8.86FLPRR286 pKa = 11.84TGSTSANKK294 pKa = 10.38LSFNATASLYY304 pKa = 8.39WHH306 pKa = 7.02EE307 pKa = 4.37KK308 pKa = 9.19
Molecular weight: 34.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
632 |
308 |
324 |
316.0 |
35.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.646 ± 0.35 | 1.424 ± 0.316 |
6.171 ± 1.371 | 4.589 ± 1.171 |
5.696 ± 0.581 | 8.861 ± 0.295 |
1.899 ± 0.878 | 4.589 ± 0.03 |
4.589 ± 1.111 | 5.38 ± 0.586 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.182 | 4.589 ± 0.426 |
4.114 ± 0.153 | 1.582 ± 0.257 |
8.228 ± 0.835 | 8.07 ± 0.946 |
8.07 ± 2.543 | 6.646 ± 1.02 |
2.532 ± 0.41 | 3.797 ± 0.526 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
