
Naumovozyma dairenensis (strain ATCC 10597 / BCRC 20456 / CBS 421 / NBRC 0211 / NRRL Y-12639) (Saccharomyces dairenensis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Naumovozyma; Naumovozyma dairenensis
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
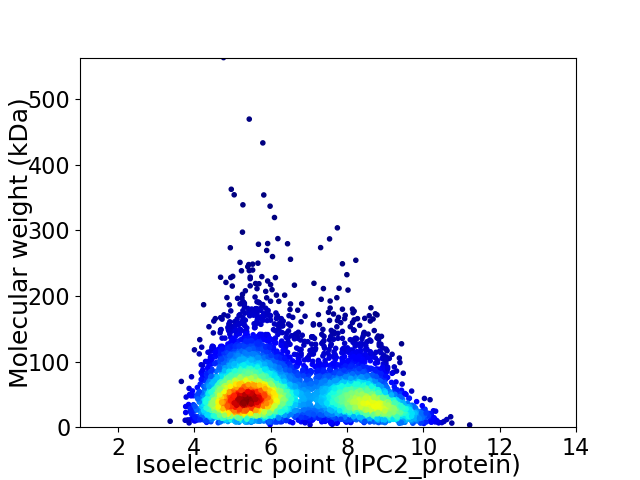
Virtual 2D-PAGE plot for 5523 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
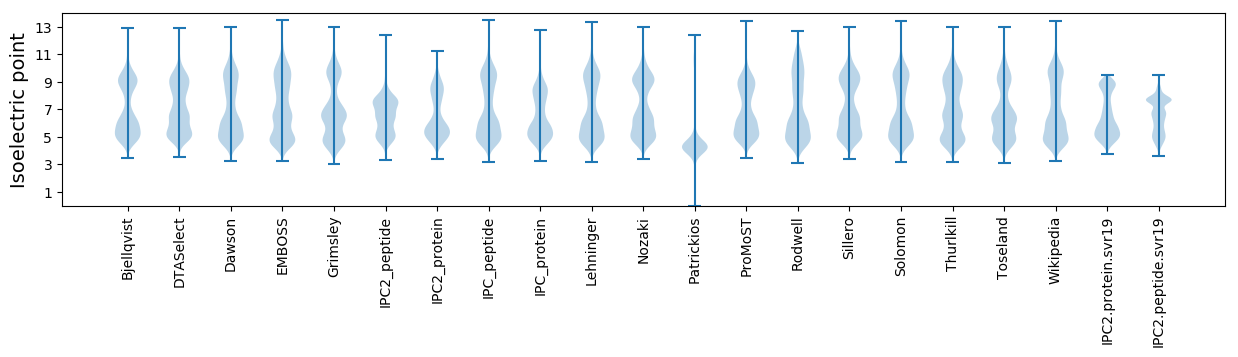
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0WG71|G0WG71_NAUDC Uncharacterized protein OS=Naumovozyma dairenensis (strain ATCC 10597 / BCRC 20456 / CBS 421 / NBRC 0211 / NRRL Y-12639) OX=1071378 GN=NDAI0I02130 PE=4 SV=1
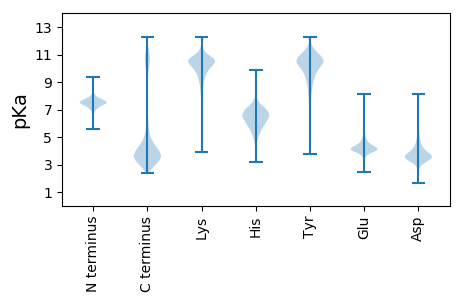
MM1 pKa = 7.6GLFDD5 pKa = 3.91QVKK8 pKa = 10.14KK9 pKa = 10.33FASGNNNNNDD19 pKa = 3.5NNNNGNDD26 pKa = 3.4NSNNNNDD33 pKa = 3.71NSNSNNDD40 pKa = 3.59DD41 pKa = 3.61FVTKK45 pKa = 10.7AEE47 pKa = 4.29NYY49 pKa = 9.25VGQDD53 pKa = 3.38KK54 pKa = 11.03VDD56 pKa = 3.41QFKK59 pKa = 11.63NKK61 pKa = 9.73IGEE64 pKa = 4.33EE65 pKa = 3.96NFNTLEE71 pKa = 4.29SKK73 pKa = 10.58VRR75 pKa = 11.84EE76 pKa = 4.21QFSKK80 pKa = 10.18TSLNDD85 pKa = 3.28KK86 pKa = 9.87NDD88 pKa = 3.51SYY90 pKa = 11.97GSSNNNGSYY99 pKa = 11.26GNSNDD104 pKa = 3.7NNNNNDD110 pKa = 3.5AYY112 pKa = 10.66GSSNNNSYY120 pKa = 11.68GNDD123 pKa = 3.29NSDD126 pKa = 4.49SYY128 pKa = 11.87GSSNNTDD135 pKa = 3.94AYY137 pKa = 10.9DD138 pKa = 3.76SSNKK142 pKa = 9.03QSSYY146 pKa = 11.84GNDD149 pKa = 3.14NDD151 pKa = 5.14DD152 pKa = 5.15SYY154 pKa = 12.24GSSNKK159 pKa = 9.44KK160 pKa = 10.04SSYY163 pKa = 11.46GNDD166 pKa = 3.11NDD168 pKa = 5.14DD169 pKa = 5.15SYY171 pKa = 12.21GSSNKK176 pKa = 9.36QSSYY180 pKa = 11.84GNDD183 pKa = 3.14NDD185 pKa = 5.14DD186 pKa = 5.15SYY188 pKa = 12.21GSSNKK193 pKa = 9.35QSSYY197 pKa = 11.69GDD199 pKa = 4.51DD200 pKa = 3.76NDD202 pKa = 5.03DD203 pKa = 4.95SYY205 pKa = 12.29GSSNKK210 pKa = 9.42KK211 pKa = 10.06SSYY214 pKa = 11.1GDD216 pKa = 3.52DD217 pKa = 3.87SYY219 pKa = 12.18GSSNKK224 pKa = 9.36QSSYY228 pKa = 11.84GNDD231 pKa = 3.14NDD233 pKa = 5.14DD234 pKa = 5.15SYY236 pKa = 12.24GSSNKK241 pKa = 9.44KK242 pKa = 10.04SSYY245 pKa = 11.46GNDD248 pKa = 3.11NDD250 pKa = 4.35DD251 pKa = 4.85SYY253 pKa = 12.13GSSHH257 pKa = 6.27KK258 pKa = 10.3QSSYY262 pKa = 11.88GNDD265 pKa = 3.14NDD267 pKa = 5.14DD268 pKa = 5.15SYY270 pKa = 12.24GSSNKK275 pKa = 9.42KK276 pKa = 10.06SSYY279 pKa = 11.1GDD281 pKa = 3.52DD282 pKa = 3.87SYY284 pKa = 12.18GSSNKK289 pKa = 9.36QSSYY293 pKa = 11.84GNDD296 pKa = 3.14NDD298 pKa = 5.14DD299 pKa = 5.15SYY301 pKa = 12.24GSSNKK306 pKa = 9.44KK307 pKa = 10.02SSYY310 pKa = 11.37GNDD313 pKa = 3.54DD314 pKa = 3.79DD315 pKa = 6.48DD316 pKa = 6.85SYY318 pKa = 12.17GSSNKK323 pKa = 9.36QSSYY327 pKa = 11.84GNDD330 pKa = 3.14NDD332 pKa = 5.14DD333 pKa = 5.15SYY335 pKa = 12.24GSSNKK340 pKa = 9.44KK341 pKa = 10.04SSYY344 pKa = 11.46GNDD347 pKa = 3.11NDD349 pKa = 5.14DD350 pKa = 5.15SYY352 pKa = 12.21GSSNKK357 pKa = 9.35QSSYY361 pKa = 11.69GDD363 pKa = 4.51DD364 pKa = 3.76NDD366 pKa = 5.03DD367 pKa = 4.43SYY369 pKa = 12.28GSSNSNTYY377 pKa = 10.61GNNNDD382 pKa = 4.21DD383 pKa = 4.53SYY385 pKa = 11.42GTDD388 pKa = 3.05RR389 pKa = 11.84NSYY392 pKa = 10.45GNSNQNSYY400 pKa = 11.9SNDD403 pKa = 3.2NNDD406 pKa = 2.72SYY408 pKa = 12.16GRR410 pKa = 11.84GNGNSGNNSSYY421 pKa = 11.56NNDD424 pKa = 3.3YY425 pKa = 11.49
MM1 pKa = 7.6GLFDD5 pKa = 3.91QVKK8 pKa = 10.14KK9 pKa = 10.33FASGNNNNNDD19 pKa = 3.5NNNNGNDD26 pKa = 3.4NSNNNNDD33 pKa = 3.71NSNSNNDD40 pKa = 3.59DD41 pKa = 3.61FVTKK45 pKa = 10.7AEE47 pKa = 4.29NYY49 pKa = 9.25VGQDD53 pKa = 3.38KK54 pKa = 11.03VDD56 pKa = 3.41QFKK59 pKa = 11.63NKK61 pKa = 9.73IGEE64 pKa = 4.33EE65 pKa = 3.96NFNTLEE71 pKa = 4.29SKK73 pKa = 10.58VRR75 pKa = 11.84EE76 pKa = 4.21QFSKK80 pKa = 10.18TSLNDD85 pKa = 3.28KK86 pKa = 9.87NDD88 pKa = 3.51SYY90 pKa = 11.97GSSNNNGSYY99 pKa = 11.26GNSNDD104 pKa = 3.7NNNNNDD110 pKa = 3.5AYY112 pKa = 10.66GSSNNNSYY120 pKa = 11.68GNDD123 pKa = 3.29NSDD126 pKa = 4.49SYY128 pKa = 11.87GSSNNTDD135 pKa = 3.94AYY137 pKa = 10.9DD138 pKa = 3.76SSNKK142 pKa = 9.03QSSYY146 pKa = 11.84GNDD149 pKa = 3.14NDD151 pKa = 5.14DD152 pKa = 5.15SYY154 pKa = 12.24GSSNKK159 pKa = 9.44KK160 pKa = 10.04SSYY163 pKa = 11.46GNDD166 pKa = 3.11NDD168 pKa = 5.14DD169 pKa = 5.15SYY171 pKa = 12.21GSSNKK176 pKa = 9.36QSSYY180 pKa = 11.84GNDD183 pKa = 3.14NDD185 pKa = 5.14DD186 pKa = 5.15SYY188 pKa = 12.21GSSNKK193 pKa = 9.35QSSYY197 pKa = 11.69GDD199 pKa = 4.51DD200 pKa = 3.76NDD202 pKa = 5.03DD203 pKa = 4.95SYY205 pKa = 12.29GSSNKK210 pKa = 9.42KK211 pKa = 10.06SSYY214 pKa = 11.1GDD216 pKa = 3.52DD217 pKa = 3.87SYY219 pKa = 12.18GSSNKK224 pKa = 9.36QSSYY228 pKa = 11.84GNDD231 pKa = 3.14NDD233 pKa = 5.14DD234 pKa = 5.15SYY236 pKa = 12.24GSSNKK241 pKa = 9.44KK242 pKa = 10.04SSYY245 pKa = 11.46GNDD248 pKa = 3.11NDD250 pKa = 4.35DD251 pKa = 4.85SYY253 pKa = 12.13GSSHH257 pKa = 6.27KK258 pKa = 10.3QSSYY262 pKa = 11.88GNDD265 pKa = 3.14NDD267 pKa = 5.14DD268 pKa = 5.15SYY270 pKa = 12.24GSSNKK275 pKa = 9.42KK276 pKa = 10.06SSYY279 pKa = 11.1GDD281 pKa = 3.52DD282 pKa = 3.87SYY284 pKa = 12.18GSSNKK289 pKa = 9.36QSSYY293 pKa = 11.84GNDD296 pKa = 3.14NDD298 pKa = 5.14DD299 pKa = 5.15SYY301 pKa = 12.24GSSNKK306 pKa = 9.44KK307 pKa = 10.02SSYY310 pKa = 11.37GNDD313 pKa = 3.54DD314 pKa = 3.79DD315 pKa = 6.48DD316 pKa = 6.85SYY318 pKa = 12.17GSSNKK323 pKa = 9.36QSSYY327 pKa = 11.84GNDD330 pKa = 3.14NDD332 pKa = 5.14DD333 pKa = 5.15SYY335 pKa = 12.24GSSNKK340 pKa = 9.44KK341 pKa = 10.04SSYY344 pKa = 11.46GNDD347 pKa = 3.11NDD349 pKa = 5.14DD350 pKa = 5.15SYY352 pKa = 12.21GSSNKK357 pKa = 9.35QSSYY361 pKa = 11.69GDD363 pKa = 4.51DD364 pKa = 3.76NDD366 pKa = 5.03DD367 pKa = 4.43SYY369 pKa = 12.28GSSNSNTYY377 pKa = 10.61GNNNDD382 pKa = 4.21DD383 pKa = 4.53SYY385 pKa = 11.42GTDD388 pKa = 3.05RR389 pKa = 11.84NSYY392 pKa = 10.45GNSNQNSYY400 pKa = 11.9SNDD403 pKa = 3.2NNDD406 pKa = 2.72SYY408 pKa = 12.16GRR410 pKa = 11.84GNGNSGNNSSYY421 pKa = 11.56NNDD424 pKa = 3.3YY425 pKa = 11.49
Molecular weight: 45.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0W3U2|G0W3U2_NAUDC Tr-type G domain-containing protein OS=Naumovozyma dairenensis (strain ATCC 10597 / BCRC 20456 / CBS 421 / NBRC 0211 / NRRL Y-12639) OX=1071378 GN=NDAI0A03230 PE=4 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.11WRR6 pKa = 11.84KK7 pKa = 9.07KK8 pKa = 6.93RR9 pKa = 11.84TRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.48RR15 pKa = 11.84KK16 pKa = 8.41RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.11WRR6 pKa = 11.84KK7 pKa = 9.07KK8 pKa = 6.93RR9 pKa = 11.84TRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.48RR15 pKa = 11.84KK16 pKa = 8.41RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
Molecular weight: 3.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2865626 |
25 |
4961 |
518.9 |
58.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.804 ± 0.028 | 1.13 ± 0.011 |
6.001 ± 0.023 | 6.562 ± 0.031 |
4.309 ± 0.021 | 4.651 ± 0.028 |
2.123 ± 0.012 | 7.229 ± 0.021 |
7.49 ± 0.032 | 9.421 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.09 ± 0.01 | 7.178 ± 0.047 |
4.313 ± 0.022 | 4.051 ± 0.028 |
4.119 ± 0.021 | 8.845 ± 0.036 |
6.326 ± 0.028 | 5.05 ± 0.023 |
0.965 ± 0.009 | 3.338 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
