
Beihai hepe-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
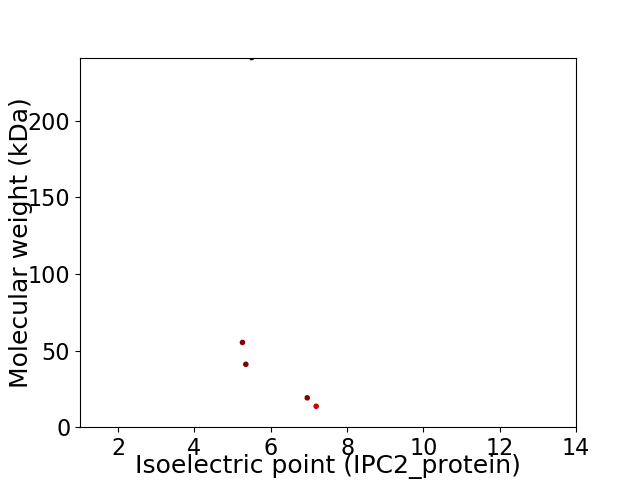
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
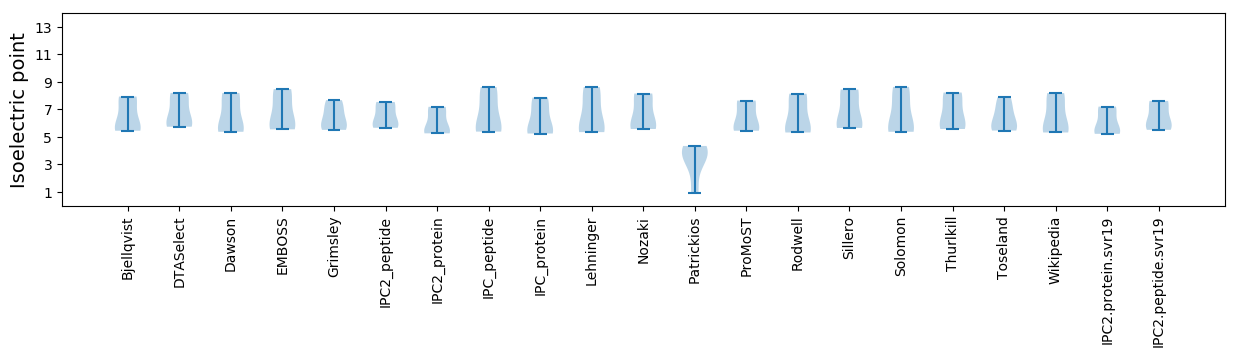
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJN7|A0A1L3KJN7_9VIRU Uncharacterized protein OS=Beihai hepe-like virus 9 OX=1922386 PE=4 SV=1
MM1 pKa = 7.41YY2 pKa = 10.88SLILLFLAGASAQTSVEE19 pKa = 4.11LIGWQFEE26 pKa = 4.25EE27 pKa = 5.09LIKK30 pKa = 10.95ASSSSNTPKK39 pKa = 10.3VVPFDD44 pKa = 4.38CNNFPNNAPYY54 pKa = 9.87WSCSDD59 pKa = 3.4TYY61 pKa = 10.59RR62 pKa = 11.84TVTLNLLIPKK72 pKa = 9.57GAYY75 pKa = 9.66LPSFVPGPTPSTDD88 pKa = 3.09DD89 pKa = 2.88MGYY92 pKa = 10.5CVGTSTIGPFPMNNMLAVMKK112 pKa = 10.21QLATIIIHH120 pKa = 5.69YY121 pKa = 8.92KK122 pKa = 8.58SHH124 pKa = 7.34FIVEE128 pKa = 4.5LQTPDD133 pKa = 4.11NYY135 pKa = 11.29DD136 pKa = 3.64FYY138 pKa = 11.28NGSPLVVAHH147 pKa = 5.36YY148 pKa = 8.94TGKK151 pKa = 9.73PLDD154 pKa = 3.64VTVSLKK160 pKa = 10.74EE161 pKa = 3.96VDD163 pKa = 3.49NMLYY167 pKa = 10.66ALSQLAFKK175 pKa = 9.01QTQFHH180 pKa = 6.04VDD182 pKa = 3.32EE183 pKa = 5.09HH184 pKa = 5.34VTAIDD189 pKa = 3.66LTGLALATGSKK200 pKa = 9.93AYY202 pKa = 10.58SSGTSVPSIIKK213 pKa = 9.27FFNYY217 pKa = 9.81TYY219 pKa = 11.11VSPFDD224 pKa = 3.99KK225 pKa = 10.79NITIISPDD233 pKa = 3.12YY234 pKa = 8.69YY235 pKa = 9.72YY236 pKa = 10.85TVFFYY241 pKa = 10.83DD242 pKa = 4.39PSAQNSISAITYY254 pKa = 7.19TNPGLQLVTEE264 pKa = 4.55LVEE267 pKa = 3.94EE268 pKa = 4.49TEE270 pKa = 3.9ISKK273 pKa = 10.71NIQFRR278 pKa = 11.84PGITVAHH285 pKa = 6.0RR286 pKa = 11.84QLMKK290 pKa = 10.61NVPIANYY297 pKa = 9.76GKK299 pKa = 10.22RR300 pKa = 11.84LTPIISFFHH309 pKa = 6.24SEE311 pKa = 3.94NYY313 pKa = 10.23LLSSWLRR320 pKa = 11.84FQNYY324 pKa = 6.34YY325 pKa = 8.71TYY327 pKa = 10.83KK328 pKa = 10.42GLNITSDD335 pKa = 3.65QLSQSIPDD343 pKa = 3.37SMRR346 pKa = 11.84NLVYY350 pKa = 9.56VQCNEE355 pKa = 3.94RR356 pKa = 11.84PDD358 pKa = 3.44MCSCKK363 pKa = 10.5GNYY366 pKa = 9.33SSQAMAIGALGYY378 pKa = 10.79DD379 pKa = 3.74SIYY382 pKa = 8.86TKK384 pKa = 10.89RR385 pKa = 11.84MNTVIYY391 pKa = 9.58PGEE394 pKa = 4.2KK395 pKa = 9.62FHH397 pKa = 5.93VHH399 pKa = 5.53SPIMSHH405 pKa = 6.05VGSVPKK411 pKa = 10.21QGRR414 pKa = 11.84YY415 pKa = 7.2QFMDD419 pKa = 3.25TTLPNLPDD427 pKa = 4.64FKK429 pKa = 11.06AVNIPSWLPEE439 pKa = 4.04SLGEE443 pKa = 4.22EE444 pKa = 4.05INGVLDD450 pKa = 3.89AVQNIYY456 pKa = 9.09EE457 pKa = 4.58HH458 pKa = 6.86IQTFWEE464 pKa = 4.05ILQHH468 pKa = 7.12IEE470 pKa = 4.23EE471 pKa = 5.51IITAIQDD478 pKa = 3.31ITSLKK483 pKa = 7.97PTGGAPPEE491 pKa = 4.23AVPVV495 pKa = 3.65
MM1 pKa = 7.41YY2 pKa = 10.88SLILLFLAGASAQTSVEE19 pKa = 4.11LIGWQFEE26 pKa = 4.25EE27 pKa = 5.09LIKK30 pKa = 10.95ASSSSNTPKK39 pKa = 10.3VVPFDD44 pKa = 4.38CNNFPNNAPYY54 pKa = 9.87WSCSDD59 pKa = 3.4TYY61 pKa = 10.59RR62 pKa = 11.84TVTLNLLIPKK72 pKa = 9.57GAYY75 pKa = 9.66LPSFVPGPTPSTDD88 pKa = 3.09DD89 pKa = 2.88MGYY92 pKa = 10.5CVGTSTIGPFPMNNMLAVMKK112 pKa = 10.21QLATIIIHH120 pKa = 5.69YY121 pKa = 8.92KK122 pKa = 8.58SHH124 pKa = 7.34FIVEE128 pKa = 4.5LQTPDD133 pKa = 4.11NYY135 pKa = 11.29DD136 pKa = 3.64FYY138 pKa = 11.28NGSPLVVAHH147 pKa = 5.36YY148 pKa = 8.94TGKK151 pKa = 9.73PLDD154 pKa = 3.64VTVSLKK160 pKa = 10.74EE161 pKa = 3.96VDD163 pKa = 3.49NMLYY167 pKa = 10.66ALSQLAFKK175 pKa = 9.01QTQFHH180 pKa = 6.04VDD182 pKa = 3.32EE183 pKa = 5.09HH184 pKa = 5.34VTAIDD189 pKa = 3.66LTGLALATGSKK200 pKa = 9.93AYY202 pKa = 10.58SSGTSVPSIIKK213 pKa = 9.27FFNYY217 pKa = 9.81TYY219 pKa = 11.11VSPFDD224 pKa = 3.99KK225 pKa = 10.79NITIISPDD233 pKa = 3.12YY234 pKa = 8.69YY235 pKa = 9.72YY236 pKa = 10.85TVFFYY241 pKa = 10.83DD242 pKa = 4.39PSAQNSISAITYY254 pKa = 7.19TNPGLQLVTEE264 pKa = 4.55LVEE267 pKa = 3.94EE268 pKa = 4.49TEE270 pKa = 3.9ISKK273 pKa = 10.71NIQFRR278 pKa = 11.84PGITVAHH285 pKa = 6.0RR286 pKa = 11.84QLMKK290 pKa = 10.61NVPIANYY297 pKa = 9.76GKK299 pKa = 10.22RR300 pKa = 11.84LTPIISFFHH309 pKa = 6.24SEE311 pKa = 3.94NYY313 pKa = 10.23LLSSWLRR320 pKa = 11.84FQNYY324 pKa = 6.34YY325 pKa = 8.71TYY327 pKa = 10.83KK328 pKa = 10.42GLNITSDD335 pKa = 3.65QLSQSIPDD343 pKa = 3.37SMRR346 pKa = 11.84NLVYY350 pKa = 9.56VQCNEE355 pKa = 3.94RR356 pKa = 11.84PDD358 pKa = 3.44MCSCKK363 pKa = 10.5GNYY366 pKa = 9.33SSQAMAIGALGYY378 pKa = 10.79DD379 pKa = 3.74SIYY382 pKa = 8.86TKK384 pKa = 10.89RR385 pKa = 11.84MNTVIYY391 pKa = 9.58PGEE394 pKa = 4.2KK395 pKa = 9.62FHH397 pKa = 5.93VHH399 pKa = 5.53SPIMSHH405 pKa = 6.05VGSVPKK411 pKa = 10.21QGRR414 pKa = 11.84YY415 pKa = 7.2QFMDD419 pKa = 3.25TTLPNLPDD427 pKa = 4.64FKK429 pKa = 11.06AVNIPSWLPEE439 pKa = 4.04SLGEE443 pKa = 4.22EE444 pKa = 4.05INGVLDD450 pKa = 3.89AVQNIYY456 pKa = 9.09EE457 pKa = 4.58HH458 pKa = 6.86IQTFWEE464 pKa = 4.05ILQHH468 pKa = 7.12IEE470 pKa = 4.23EE471 pKa = 5.51IITAIQDD478 pKa = 3.31ITSLKK483 pKa = 7.97PTGGAPPEE491 pKa = 4.23AVPVV495 pKa = 3.65
Molecular weight: 55.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJP0|A0A1L3KJP0_9VIRU Uncharacterized protein OS=Beihai hepe-like virus 9 OX=1922386 PE=4 SV=1
MM1 pKa = 6.65THH3 pKa = 6.72ILAFRR8 pKa = 11.84VSNFVFQVEE17 pKa = 4.35FDD19 pKa = 3.86YY20 pKa = 11.61DD21 pKa = 3.97DD22 pKa = 4.78APLEE26 pKa = 4.06ARR28 pKa = 11.84KK29 pKa = 9.36QLISLGNPKK38 pKa = 10.09GRR40 pKa = 11.84GIPEE44 pKa = 4.23PLHH47 pKa = 6.01VVEE50 pKa = 4.58YY51 pKa = 11.15SRR53 pKa = 11.84FEE55 pKa = 3.86GGKK58 pKa = 9.99VIEE61 pKa = 4.36VEE63 pKa = 3.93PNYY66 pKa = 10.22ILYY69 pKa = 10.05HH70 pKa = 5.78KK71 pKa = 10.54QPLHH75 pKa = 6.25PFFVSKK81 pKa = 10.24FRR83 pKa = 11.84NRR85 pKa = 11.84LNFVRR90 pKa = 11.84VISNCSQQSHH100 pKa = 5.92IPEE103 pKa = 3.98SFYY106 pKa = 11.19TNFISNWQMQQ116 pKa = 3.28
MM1 pKa = 6.65THH3 pKa = 6.72ILAFRR8 pKa = 11.84VSNFVFQVEE17 pKa = 4.35FDD19 pKa = 3.86YY20 pKa = 11.61DD21 pKa = 3.97DD22 pKa = 4.78APLEE26 pKa = 4.06ARR28 pKa = 11.84KK29 pKa = 9.36QLISLGNPKK38 pKa = 10.09GRR40 pKa = 11.84GIPEE44 pKa = 4.23PLHH47 pKa = 6.01VVEE50 pKa = 4.58YY51 pKa = 11.15SRR53 pKa = 11.84FEE55 pKa = 3.86GGKK58 pKa = 9.99VIEE61 pKa = 4.36VEE63 pKa = 3.93PNYY66 pKa = 10.22ILYY69 pKa = 10.05HH70 pKa = 5.78KK71 pKa = 10.54QPLHH75 pKa = 6.25PFFVSKK81 pKa = 10.24FRR83 pKa = 11.84NRR85 pKa = 11.84LNFVRR90 pKa = 11.84VISNCSQQSHH100 pKa = 5.92IPEE103 pKa = 3.98SFYY106 pKa = 11.19TNFISNWQMQQ116 pKa = 3.28
Molecular weight: 13.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3302 |
116 |
2150 |
660.4 |
74.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.389 ± 0.873 | 1.575 ± 0.26 |
5.451 ± 0.665 | 5.996 ± 0.712 |
4.21 ± 0.643 | 5.996 ± 0.286 |
3.331 ± 0.423 | 6.299 ± 0.714 |
5.451 ± 0.488 | 7.117 ± 0.522 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.271 ± 0.24 | 4.815 ± 0.435 |
5.27 ± 0.601 | 4.573 ± 0.203 |
3.786 ± 0.609 | 6.602 ± 1.024 |
7.268 ± 0.429 | 6.784 ± 0.288 |
1.211 ± 0.083 | 4.603 ± 0.537 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
