
Sanxia tombus-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.24
Get precalculated fractions of proteins
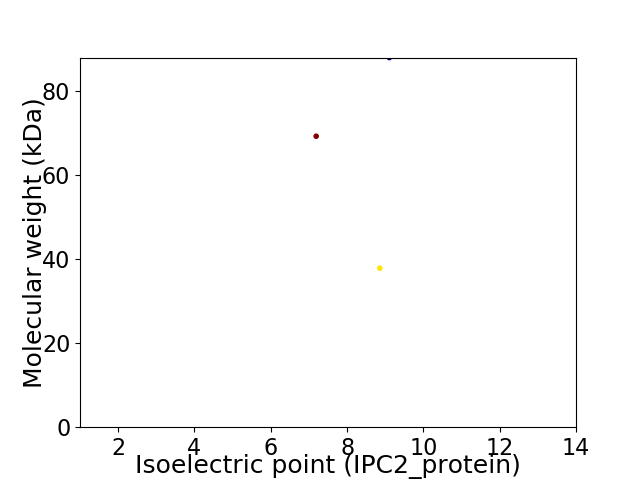
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGB8|A0A1L3KGB8_9VIRU Uncharacterized protein OS=Sanxia tombus-like virus 9 OX=1923393 PE=4 SV=1
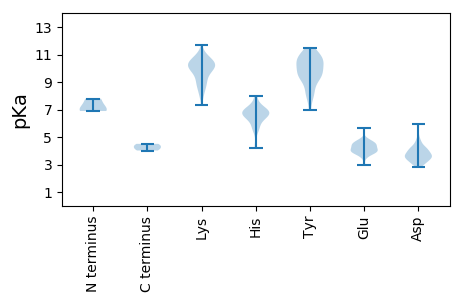
MM1 pKa = 7.79LSRR4 pKa = 11.84LHH6 pKa = 6.34VCFHH10 pKa = 6.05HH11 pKa = 7.42RR12 pKa = 11.84GRR14 pKa = 11.84QLLPPLCSLVPMVPAAHH31 pKa = 6.54VDD33 pKa = 3.63NVVSSIEE40 pKa = 3.78LRR42 pKa = 11.84VARR45 pKa = 11.84EE46 pKa = 3.86VPRR49 pKa = 11.84PHH51 pKa = 7.32PLVCAPKK58 pKa = 10.45LRR60 pKa = 11.84GKK62 pKa = 9.92LRR64 pKa = 11.84PYY66 pKa = 10.19LLHH69 pKa = 6.95PAPLVPTPLAMPLSPPRR86 pKa = 11.84KK87 pKa = 8.44VPPRR91 pKa = 11.84KK92 pKa = 8.64PQHH95 pKa = 6.53LSSQLWEE102 pKa = 4.21KK103 pKa = 10.29VQAAVNPEE111 pKa = 4.06LVLATTANATDD122 pKa = 3.63TSLPTAAADD131 pKa = 3.58HH132 pKa = 6.59RR133 pKa = 11.84LDD135 pKa = 4.02PSSQAVHH142 pKa = 6.86PCRR145 pKa = 11.84NPEE148 pKa = 3.91NCSLLLFPGVPKK160 pKa = 10.42SDD162 pKa = 3.27PHH164 pKa = 7.99GPEE167 pKa = 3.79SLAMVQKK174 pKa = 11.03AMGMVPEE181 pKa = 4.54SRR183 pKa = 11.84TEE185 pKa = 3.9NLQQLADD192 pKa = 4.09EE193 pKa = 4.5YY194 pKa = 10.79RR195 pKa = 11.84ANHH198 pKa = 6.83LLPVLGRR205 pKa = 11.84VHH207 pKa = 7.08PLPWEE212 pKa = 3.74EE213 pKa = 3.51WLQRR217 pKa = 11.84FPSNRR222 pKa = 11.84SLQLNDD228 pKa = 3.15ARR230 pKa = 11.84SNVQLRR236 pKa = 11.84GLLKK240 pKa = 10.3SDD242 pKa = 4.03CKK244 pKa = 10.89VDD246 pKa = 3.69CFLKK250 pKa = 11.06VEE252 pKa = 4.36TSTNATDD259 pKa = 3.86PRR261 pKa = 11.84NISPRR266 pKa = 11.84SAEE269 pKa = 3.98FLSVLGPYY277 pKa = 9.49VAGLEE282 pKa = 4.16NKK284 pKa = 9.3IRR286 pKa = 11.84TLHH289 pKa = 6.66DD290 pKa = 3.75HH291 pKa = 6.95DD292 pKa = 4.65QIPYY296 pKa = 9.23LVKK299 pKa = 10.64GLTPSARR306 pKa = 11.84AQIVRR311 pKa = 11.84RR312 pKa = 11.84KK313 pKa = 7.88EE314 pKa = 3.83RR315 pKa = 11.84AWVVEE320 pKa = 3.85IDD322 pKa = 3.43YY323 pKa = 11.06SRR325 pKa = 11.84YY326 pKa = 11.07DD327 pKa = 3.31MTISEE332 pKa = 4.2DD333 pKa = 3.62VIRR336 pKa = 11.84EE337 pKa = 4.03LEE339 pKa = 4.04IKK341 pKa = 10.16NFKK344 pKa = 10.69ALFVSDD350 pKa = 4.58EE351 pKa = 4.4DD352 pKa = 4.1PALDD356 pKa = 3.79AVLDD360 pKa = 4.23ALISMNGYY368 pKa = 9.72NSLGVKK374 pKa = 8.64YY375 pKa = 8.77TVDD378 pKa = 3.43GTRR381 pKa = 11.84ASGDD385 pKa = 2.95AHH387 pKa = 5.71TSIGNGLLGRR397 pKa = 11.84FLLWVCLRR405 pKa = 11.84GLPDD409 pKa = 3.75TVWDD413 pKa = 4.21SVHH416 pKa = 6.61EE417 pKa = 4.52GDD419 pKa = 5.97DD420 pKa = 3.52GLLFVDD426 pKa = 5.8DD427 pKa = 4.61GWQHH431 pKa = 6.84LVMHH435 pKa = 7.05CLQVLPCFGFQAKK448 pKa = 9.66LRR450 pKa = 11.84LCRR453 pKa = 11.84LDD455 pKa = 4.93DD456 pKa = 4.1ALFCGRR462 pKa = 11.84LNVNTSNGAIEE473 pKa = 4.24IADD476 pKa = 3.69IPRR479 pKa = 11.84TLGKK483 pKa = 10.21FHH485 pKa = 6.92TSCKK489 pKa = 9.83TGDD492 pKa = 3.54ARR494 pKa = 11.84TLALAKK500 pKa = 10.35AFSYY504 pKa = 10.78YY505 pKa = 9.38HH506 pKa = 6.69TDD508 pKa = 2.88RR509 pKa = 11.84DD510 pKa = 4.1TPIIGPLCWALIQHH524 pKa = 6.64LRR526 pKa = 11.84PLVSPRR532 pKa = 11.84AFTRR536 pKa = 11.84RR537 pKa = 11.84INTEE541 pKa = 2.94KK542 pKa = 9.78WFEE545 pKa = 4.04RR546 pKa = 11.84EE547 pKa = 3.88KK548 pKa = 10.74IEE550 pKa = 5.64AGMKK554 pKa = 7.9VQRR557 pKa = 11.84PPRR560 pKa = 11.84ITPEE564 pKa = 3.02ARR566 pKa = 11.84AAVDD570 pKa = 3.33RR571 pKa = 11.84VFDD574 pKa = 3.55VGYY577 pKa = 10.67GSQLVLEE584 pKa = 4.79KK585 pKa = 10.64QLSSWASGVSEE596 pKa = 4.56VPPLILTGEE605 pKa = 4.32EE606 pKa = 4.54VVDD609 pKa = 3.57HH610 pKa = 6.69TDD612 pKa = 3.06GVKK615 pKa = 9.55TVTVYY620 pKa = 11.17AA621 pKa = 4.5
MM1 pKa = 7.79LSRR4 pKa = 11.84LHH6 pKa = 6.34VCFHH10 pKa = 6.05HH11 pKa = 7.42RR12 pKa = 11.84GRR14 pKa = 11.84QLLPPLCSLVPMVPAAHH31 pKa = 6.54VDD33 pKa = 3.63NVVSSIEE40 pKa = 3.78LRR42 pKa = 11.84VARR45 pKa = 11.84EE46 pKa = 3.86VPRR49 pKa = 11.84PHH51 pKa = 7.32PLVCAPKK58 pKa = 10.45LRR60 pKa = 11.84GKK62 pKa = 9.92LRR64 pKa = 11.84PYY66 pKa = 10.19LLHH69 pKa = 6.95PAPLVPTPLAMPLSPPRR86 pKa = 11.84KK87 pKa = 8.44VPPRR91 pKa = 11.84KK92 pKa = 8.64PQHH95 pKa = 6.53LSSQLWEE102 pKa = 4.21KK103 pKa = 10.29VQAAVNPEE111 pKa = 4.06LVLATTANATDD122 pKa = 3.63TSLPTAAADD131 pKa = 3.58HH132 pKa = 6.59RR133 pKa = 11.84LDD135 pKa = 4.02PSSQAVHH142 pKa = 6.86PCRR145 pKa = 11.84NPEE148 pKa = 3.91NCSLLLFPGVPKK160 pKa = 10.42SDD162 pKa = 3.27PHH164 pKa = 7.99GPEE167 pKa = 3.79SLAMVQKK174 pKa = 11.03AMGMVPEE181 pKa = 4.54SRR183 pKa = 11.84TEE185 pKa = 3.9NLQQLADD192 pKa = 4.09EE193 pKa = 4.5YY194 pKa = 10.79RR195 pKa = 11.84ANHH198 pKa = 6.83LLPVLGRR205 pKa = 11.84VHH207 pKa = 7.08PLPWEE212 pKa = 3.74EE213 pKa = 3.51WLQRR217 pKa = 11.84FPSNRR222 pKa = 11.84SLQLNDD228 pKa = 3.15ARR230 pKa = 11.84SNVQLRR236 pKa = 11.84GLLKK240 pKa = 10.3SDD242 pKa = 4.03CKK244 pKa = 10.89VDD246 pKa = 3.69CFLKK250 pKa = 11.06VEE252 pKa = 4.36TSTNATDD259 pKa = 3.86PRR261 pKa = 11.84NISPRR266 pKa = 11.84SAEE269 pKa = 3.98FLSVLGPYY277 pKa = 9.49VAGLEE282 pKa = 4.16NKK284 pKa = 9.3IRR286 pKa = 11.84TLHH289 pKa = 6.66DD290 pKa = 3.75HH291 pKa = 6.95DD292 pKa = 4.65QIPYY296 pKa = 9.23LVKK299 pKa = 10.64GLTPSARR306 pKa = 11.84AQIVRR311 pKa = 11.84RR312 pKa = 11.84KK313 pKa = 7.88EE314 pKa = 3.83RR315 pKa = 11.84AWVVEE320 pKa = 3.85IDD322 pKa = 3.43YY323 pKa = 11.06SRR325 pKa = 11.84YY326 pKa = 11.07DD327 pKa = 3.31MTISEE332 pKa = 4.2DD333 pKa = 3.62VIRR336 pKa = 11.84EE337 pKa = 4.03LEE339 pKa = 4.04IKK341 pKa = 10.16NFKK344 pKa = 10.69ALFVSDD350 pKa = 4.58EE351 pKa = 4.4DD352 pKa = 4.1PALDD356 pKa = 3.79AVLDD360 pKa = 4.23ALISMNGYY368 pKa = 9.72NSLGVKK374 pKa = 8.64YY375 pKa = 8.77TVDD378 pKa = 3.43GTRR381 pKa = 11.84ASGDD385 pKa = 2.95AHH387 pKa = 5.71TSIGNGLLGRR397 pKa = 11.84FLLWVCLRR405 pKa = 11.84GLPDD409 pKa = 3.75TVWDD413 pKa = 4.21SVHH416 pKa = 6.61EE417 pKa = 4.52GDD419 pKa = 5.97DD420 pKa = 3.52GLLFVDD426 pKa = 5.8DD427 pKa = 4.61GWQHH431 pKa = 6.84LVMHH435 pKa = 7.05CLQVLPCFGFQAKK448 pKa = 9.66LRR450 pKa = 11.84LCRR453 pKa = 11.84LDD455 pKa = 4.93DD456 pKa = 4.1ALFCGRR462 pKa = 11.84LNVNTSNGAIEE473 pKa = 4.24IADD476 pKa = 3.69IPRR479 pKa = 11.84TLGKK483 pKa = 10.21FHH485 pKa = 6.92TSCKK489 pKa = 9.83TGDD492 pKa = 3.54ARR494 pKa = 11.84TLALAKK500 pKa = 10.35AFSYY504 pKa = 10.78YY505 pKa = 9.38HH506 pKa = 6.69TDD508 pKa = 2.88RR509 pKa = 11.84DD510 pKa = 4.1TPIIGPLCWALIQHH524 pKa = 6.64LRR526 pKa = 11.84PLVSPRR532 pKa = 11.84AFTRR536 pKa = 11.84RR537 pKa = 11.84INTEE541 pKa = 2.94KK542 pKa = 9.78WFEE545 pKa = 4.04RR546 pKa = 11.84EE547 pKa = 3.88KK548 pKa = 10.74IEE550 pKa = 5.64AGMKK554 pKa = 7.9VQRR557 pKa = 11.84PPRR560 pKa = 11.84ITPEE564 pKa = 3.02ARR566 pKa = 11.84AAVDD570 pKa = 3.33RR571 pKa = 11.84VFDD574 pKa = 3.55VGYY577 pKa = 10.67GSQLVLEE584 pKa = 4.79KK585 pKa = 10.64QLSSWASGVSEE596 pKa = 4.56VPPLILTGEE605 pKa = 4.32EE606 pKa = 4.54VVDD609 pKa = 3.57HH610 pKa = 6.69TDD612 pKa = 3.06GVKK615 pKa = 9.55TVTVYY620 pKa = 11.17AA621 pKa = 4.5
Molecular weight: 69.14 kDa
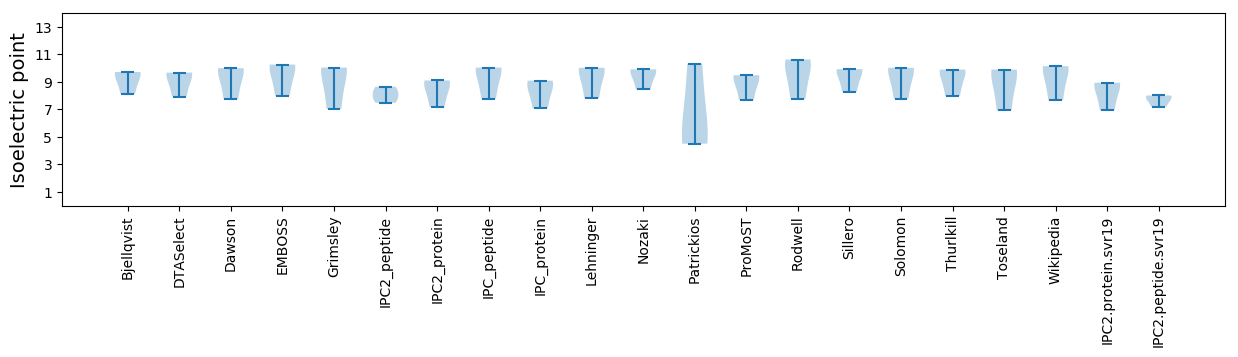
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGI7|A0A1L3KGI7_9VIRU Chropara_Vmeth domain-containing protein OS=Sanxia tombus-like virus 9 OX=1923393 PE=4 SV=1
MM1 pKa = 6.89VAWLPVVIGFAITALMAPRR20 pKa = 11.84IEE22 pKa = 4.58EE23 pKa = 4.0DD24 pKa = 2.98LSYY27 pKa = 11.16YY28 pKa = 10.55EE29 pKa = 4.85SLGAPGWRR37 pKa = 11.84TVDD40 pKa = 3.1AYY42 pKa = 11.47LPILKK47 pKa = 10.23YY48 pKa = 10.68LDD50 pKa = 4.27LVPSPEE56 pKa = 3.79RR57 pKa = 11.84LEE59 pKa = 4.43RR60 pKa = 11.84LRR62 pKa = 11.84LDD64 pKa = 3.48CAIIQGEE71 pKa = 4.0IHH73 pKa = 5.9EE74 pKa = 4.48WEE76 pKa = 3.5WWAEE80 pKa = 3.95YY81 pKa = 10.02FPTMAGEE88 pKa = 4.5RR89 pKa = 11.84PALPYY94 pKa = 9.83RR95 pKa = 11.84CSYY98 pKa = 9.35FVWNRR103 pKa = 11.84TAKK106 pKa = 10.06PAVTSRR112 pKa = 11.84LDD114 pKa = 3.81LLMSAYY120 pKa = 9.17ATIAKK125 pKa = 9.98GLLFLGAWVWINVALIVLAFICCLLAGVLLVVKK158 pKa = 10.29VYY160 pKa = 8.38LTYY163 pKa = 10.43YY164 pKa = 9.3RR165 pKa = 11.84GVRR168 pKa = 11.84VRR170 pKa = 11.84APTPVLGKK178 pKa = 10.52NEE180 pKa = 3.94VRR182 pKa = 11.84DD183 pKa = 3.81LLAVEE188 pKa = 4.83LSKK191 pKa = 10.16TPIDD195 pKa = 4.74KK196 pKa = 10.53IQLLADD202 pKa = 4.35GKK204 pKa = 9.91GHH206 pKa = 6.87PVLAKK211 pKa = 9.84TRR213 pKa = 11.84ATLEE217 pKa = 4.23TACQQVLLRR226 pKa = 11.84MNNKK230 pKa = 8.94IRR232 pKa = 11.84DD233 pKa = 3.72VGGSVSRR240 pKa = 11.84NQKK243 pKa = 9.62LGKK246 pKa = 9.34SLHH249 pKa = 5.63VCFPVLTAADD259 pKa = 4.0RR260 pKa = 11.84ARR262 pKa = 11.84VNRR265 pKa = 11.84ALANDD270 pKa = 3.52VGFHH274 pKa = 6.89LGHH277 pKa = 6.37EE278 pKa = 4.75CDD280 pKa = 3.67VKK282 pKa = 11.01HH283 pKa = 6.47IPTYY287 pKa = 9.77MSYY290 pKa = 10.86TDD292 pKa = 3.64FHH294 pKa = 6.94MPLDD298 pKa = 3.65MLVRR302 pKa = 11.84TVTSPTLIITHH313 pKa = 6.78DD314 pKa = 4.07FRR316 pKa = 11.84NMPAKK321 pKa = 10.62LFDD324 pKa = 3.89EE325 pKa = 4.65EE326 pKa = 4.37ATISVIGGLVNMTTRR341 pKa = 11.84GGSKK345 pKa = 8.78YY346 pKa = 7.0THH348 pKa = 7.27GYY350 pKa = 9.56HH351 pKa = 6.34SWDD354 pKa = 3.77DD355 pKa = 3.64EE356 pKa = 4.78GSVVTKK362 pKa = 10.65SGAFYY367 pKa = 8.01YY368 pKa = 10.63HH369 pKa = 6.96KK370 pKa = 10.54VAEE373 pKa = 4.77LGDD376 pKa = 4.31SIVIYY381 pKa = 8.07ATPLNGDD388 pKa = 3.87HH389 pKa = 7.84DD390 pKa = 4.14PTQDD394 pKa = 3.5NEE396 pKa = 4.28LQSSVNCYY404 pKa = 9.59TGIKK408 pKa = 9.08YY409 pKa = 10.39RR410 pKa = 11.84GGRR413 pKa = 11.84AEE415 pKa = 4.1LHH417 pKa = 5.46NGEE420 pKa = 4.66YY421 pKa = 10.3IMRR424 pKa = 11.84DD425 pKa = 3.23DD426 pKa = 4.03QGNLRR431 pKa = 11.84GAVSARR437 pKa = 11.84TVSRR441 pKa = 11.84VAHH444 pKa = 5.42SFGYY448 pKa = 10.46AKK450 pKa = 10.12RR451 pKa = 11.84DD452 pKa = 3.47AKK454 pKa = 11.08FEE456 pKa = 4.15TGLAANLRR464 pKa = 11.84ARR466 pKa = 11.84FTQDD470 pKa = 3.69GEE472 pKa = 4.36DD473 pKa = 3.49HH474 pKa = 6.74SLLPEE479 pKa = 4.18ATNLTVALADD489 pKa = 5.4RR490 pKa = 11.84IALAADD496 pKa = 4.25MSAVSCPTEE505 pKa = 3.64MNFFQRR511 pKa = 11.84VMWNAYY517 pKa = 7.91FALVSVLPTSLQLARR532 pKa = 11.84TVRR535 pKa = 11.84QMLLQSEE542 pKa = 4.6HH543 pKa = 4.24VRR545 pKa = 11.84FRR547 pKa = 11.84KK548 pKa = 9.1WMWEE552 pKa = 3.87EE553 pKa = 3.59VHH555 pKa = 6.46IPNYY559 pKa = 10.21LVSTPGLVSLRR570 pKa = 11.84ADD572 pKa = 3.11AVKK575 pKa = 10.53ASRR578 pKa = 11.84LFPSPGAATAAPAVLACTNGAGCACGQCGQQHH610 pKa = 5.67RR611 pKa = 11.84TQGSSGSPAAASLSLRR627 pKa = 11.84PQTTRR632 pKa = 11.84QTTSLSPTPSPTSTNTTSNAPVPAKK657 pKa = 10.46KK658 pKa = 9.1GTASQAPAPQQPAVGKK674 pKa = 9.91GAGSGKK680 pKa = 10.33SRR682 pKa = 11.84IGPCNYY688 pKa = 9.51CKK690 pKa = 10.34RR691 pKa = 11.84HH692 pKa = 4.39GHH694 pKa = 6.15VIANCRR700 pKa = 11.84SRR702 pKa = 11.84PSAGSVKK709 pKa = 10.4PGGSPVQKK717 pKa = 10.02PGKK720 pKa = 8.46LQPAAVPGGAQVRR733 pKa = 11.84PSRR736 pKa = 11.84AGKK739 pKa = 9.25PSNGAKK745 pKa = 10.61GNGNGTRR752 pKa = 11.84KK753 pKa = 9.93QNGKK757 pKa = 8.73PAATRR762 pKa = 11.84GRR764 pKa = 11.84VQSKK768 pKa = 10.42SPAPGARR775 pKa = 11.84ARR777 pKa = 11.84PPPPVGRR784 pKa = 11.84VAAAVPVKK792 pKa = 10.25PFTPAQRR799 pKa = 11.84RR800 pKa = 11.84AQQCATSRR808 pKa = 11.84AVKK811 pKa = 10.08II812 pKa = 4.02
MM1 pKa = 6.89VAWLPVVIGFAITALMAPRR20 pKa = 11.84IEE22 pKa = 4.58EE23 pKa = 4.0DD24 pKa = 2.98LSYY27 pKa = 11.16YY28 pKa = 10.55EE29 pKa = 4.85SLGAPGWRR37 pKa = 11.84TVDD40 pKa = 3.1AYY42 pKa = 11.47LPILKK47 pKa = 10.23YY48 pKa = 10.68LDD50 pKa = 4.27LVPSPEE56 pKa = 3.79RR57 pKa = 11.84LEE59 pKa = 4.43RR60 pKa = 11.84LRR62 pKa = 11.84LDD64 pKa = 3.48CAIIQGEE71 pKa = 4.0IHH73 pKa = 5.9EE74 pKa = 4.48WEE76 pKa = 3.5WWAEE80 pKa = 3.95YY81 pKa = 10.02FPTMAGEE88 pKa = 4.5RR89 pKa = 11.84PALPYY94 pKa = 9.83RR95 pKa = 11.84CSYY98 pKa = 9.35FVWNRR103 pKa = 11.84TAKK106 pKa = 10.06PAVTSRR112 pKa = 11.84LDD114 pKa = 3.81LLMSAYY120 pKa = 9.17ATIAKK125 pKa = 9.98GLLFLGAWVWINVALIVLAFICCLLAGVLLVVKK158 pKa = 10.29VYY160 pKa = 8.38LTYY163 pKa = 10.43YY164 pKa = 9.3RR165 pKa = 11.84GVRR168 pKa = 11.84VRR170 pKa = 11.84APTPVLGKK178 pKa = 10.52NEE180 pKa = 3.94VRR182 pKa = 11.84DD183 pKa = 3.81LLAVEE188 pKa = 4.83LSKK191 pKa = 10.16TPIDD195 pKa = 4.74KK196 pKa = 10.53IQLLADD202 pKa = 4.35GKK204 pKa = 9.91GHH206 pKa = 6.87PVLAKK211 pKa = 9.84TRR213 pKa = 11.84ATLEE217 pKa = 4.23TACQQVLLRR226 pKa = 11.84MNNKK230 pKa = 8.94IRR232 pKa = 11.84DD233 pKa = 3.72VGGSVSRR240 pKa = 11.84NQKK243 pKa = 9.62LGKK246 pKa = 9.34SLHH249 pKa = 5.63VCFPVLTAADD259 pKa = 4.0RR260 pKa = 11.84ARR262 pKa = 11.84VNRR265 pKa = 11.84ALANDD270 pKa = 3.52VGFHH274 pKa = 6.89LGHH277 pKa = 6.37EE278 pKa = 4.75CDD280 pKa = 3.67VKK282 pKa = 11.01HH283 pKa = 6.47IPTYY287 pKa = 9.77MSYY290 pKa = 10.86TDD292 pKa = 3.64FHH294 pKa = 6.94MPLDD298 pKa = 3.65MLVRR302 pKa = 11.84TVTSPTLIITHH313 pKa = 6.78DD314 pKa = 4.07FRR316 pKa = 11.84NMPAKK321 pKa = 10.62LFDD324 pKa = 3.89EE325 pKa = 4.65EE326 pKa = 4.37ATISVIGGLVNMTTRR341 pKa = 11.84GGSKK345 pKa = 8.78YY346 pKa = 7.0THH348 pKa = 7.27GYY350 pKa = 9.56HH351 pKa = 6.34SWDD354 pKa = 3.77DD355 pKa = 3.64EE356 pKa = 4.78GSVVTKK362 pKa = 10.65SGAFYY367 pKa = 8.01YY368 pKa = 10.63HH369 pKa = 6.96KK370 pKa = 10.54VAEE373 pKa = 4.77LGDD376 pKa = 4.31SIVIYY381 pKa = 8.07ATPLNGDD388 pKa = 3.87HH389 pKa = 7.84DD390 pKa = 4.14PTQDD394 pKa = 3.5NEE396 pKa = 4.28LQSSVNCYY404 pKa = 9.59TGIKK408 pKa = 9.08YY409 pKa = 10.39RR410 pKa = 11.84GGRR413 pKa = 11.84AEE415 pKa = 4.1LHH417 pKa = 5.46NGEE420 pKa = 4.66YY421 pKa = 10.3IMRR424 pKa = 11.84DD425 pKa = 3.23DD426 pKa = 4.03QGNLRR431 pKa = 11.84GAVSARR437 pKa = 11.84TVSRR441 pKa = 11.84VAHH444 pKa = 5.42SFGYY448 pKa = 10.46AKK450 pKa = 10.12RR451 pKa = 11.84DD452 pKa = 3.47AKK454 pKa = 11.08FEE456 pKa = 4.15TGLAANLRR464 pKa = 11.84ARR466 pKa = 11.84FTQDD470 pKa = 3.69GEE472 pKa = 4.36DD473 pKa = 3.49HH474 pKa = 6.74SLLPEE479 pKa = 4.18ATNLTVALADD489 pKa = 5.4RR490 pKa = 11.84IALAADD496 pKa = 4.25MSAVSCPTEE505 pKa = 3.64MNFFQRR511 pKa = 11.84VMWNAYY517 pKa = 7.91FALVSVLPTSLQLARR532 pKa = 11.84TVRR535 pKa = 11.84QMLLQSEE542 pKa = 4.6HH543 pKa = 4.24VRR545 pKa = 11.84FRR547 pKa = 11.84KK548 pKa = 9.1WMWEE552 pKa = 3.87EE553 pKa = 3.59VHH555 pKa = 6.46IPNYY559 pKa = 10.21LVSTPGLVSLRR570 pKa = 11.84ADD572 pKa = 3.11AVKK575 pKa = 10.53ASRR578 pKa = 11.84LFPSPGAATAAPAVLACTNGAGCACGQCGQQHH610 pKa = 5.67RR611 pKa = 11.84TQGSSGSPAAASLSLRR627 pKa = 11.84PQTTRR632 pKa = 11.84QTTSLSPTPSPTSTNTTSNAPVPAKK657 pKa = 10.46KK658 pKa = 9.1GTASQAPAPQQPAVGKK674 pKa = 9.91GAGSGKK680 pKa = 10.33SRR682 pKa = 11.84IGPCNYY688 pKa = 9.51CKK690 pKa = 10.34RR691 pKa = 11.84HH692 pKa = 4.39GHH694 pKa = 6.15VIANCRR700 pKa = 11.84SRR702 pKa = 11.84PSAGSVKK709 pKa = 10.4PGGSPVQKK717 pKa = 10.02PGKK720 pKa = 8.46LQPAAVPGGAQVRR733 pKa = 11.84PSRR736 pKa = 11.84AGKK739 pKa = 9.25PSNGAKK745 pKa = 10.61GNGNGTRR752 pKa = 11.84KK753 pKa = 9.93QNGKK757 pKa = 8.73PAATRR762 pKa = 11.84GRR764 pKa = 11.84VQSKK768 pKa = 10.42SPAPGARR775 pKa = 11.84ARR777 pKa = 11.84PPPPVGRR784 pKa = 11.84VAAAVPVKK792 pKa = 10.25PFTPAQRR799 pKa = 11.84RR800 pKa = 11.84AQQCATSRR808 pKa = 11.84AVKK811 pKa = 10.08II812 pKa = 4.02
Molecular weight: 87.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1787 |
354 |
812 |
595.7 |
64.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.289 ± 1.294 | 1.903 ± 0.246 |
4.98 ± 0.736 | 3.693 ± 0.59 |
2.63 ± 0.094 | 7.219 ± 0.854 |
2.462 ± 0.53 | 3.078 ± 0.32 |
4.757 ± 0.425 | 9.905 ± 1.187 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.847 ± 0.109 | 3.693 ± 0.196 |
7.499 ± 0.326 | 3.637 ± 0.192 |
6.771 ± 0.458 | 7.163 ± 0.504 |
6.715 ± 0.899 | 8.842 ± 0.549 |
1.679 ± 0.141 | 2.238 ± 0.476 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
