
Human papillomavirus 139
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 7
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
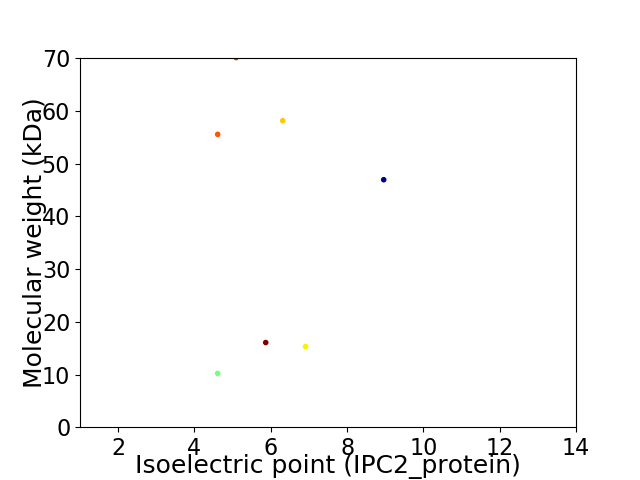
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I6MRE3|I6MRE3_9PAPI Replication protein E1 OS=Human papillomavirus 139 OX=1070412 GN=E1 PE=3 SV=1
MM1 pKa = 7.49HH2 pKa = 7.47GEE4 pKa = 4.01VPDD7 pKa = 3.68IKK9 pKa = 11.03DD10 pKa = 3.46VILEE14 pKa = 4.02NLEE17 pKa = 4.84DD18 pKa = 5.5LILPANLLSDD28 pKa = 3.99EE29 pKa = 4.76DD30 pKa = 4.19LSAEE34 pKa = 4.16AEE36 pKa = 4.23LEE38 pKa = 4.34SAHH41 pKa = 5.35VLYY44 pKa = 10.8RR45 pKa = 11.84IVTYY49 pKa = 6.89CHH51 pKa = 6.63CGTRR55 pKa = 11.84IKK57 pKa = 10.66FSVLASPEE65 pKa = 4.29GIRR68 pKa = 11.84GLEE71 pKa = 3.85LLLLEE76 pKa = 4.73EE77 pKa = 4.77VSFLCTGCSKK87 pKa = 10.94GLNNGRR93 pKa = 3.95
MM1 pKa = 7.49HH2 pKa = 7.47GEE4 pKa = 4.01VPDD7 pKa = 3.68IKK9 pKa = 11.03DD10 pKa = 3.46VILEE14 pKa = 4.02NLEE17 pKa = 4.84DD18 pKa = 5.5LILPANLLSDD28 pKa = 3.99EE29 pKa = 4.76DD30 pKa = 4.19LSAEE34 pKa = 4.16AEE36 pKa = 4.23LEE38 pKa = 4.34SAHH41 pKa = 5.35VLYY44 pKa = 10.8RR45 pKa = 11.84IVTYY49 pKa = 6.89CHH51 pKa = 6.63CGTRR55 pKa = 11.84IKK57 pKa = 10.66FSVLASPEE65 pKa = 4.29GIRR68 pKa = 11.84GLEE71 pKa = 3.85LLLLEE76 pKa = 4.73EE77 pKa = 4.77VSFLCTGCSKK87 pKa = 10.94GLNNGRR93 pKa = 3.95
Molecular weight: 10.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I6MRE5|I6MRE5_9PAPI Uncharacterized protein (Fragment) OS=Human papillomavirus 139 OX=1070412 GN=E4 PE=4 SV=1
MM1 pKa = 6.94EE2 pKa = 4.99TLNEE6 pKa = 4.24RR7 pKa = 11.84YY8 pKa = 9.3DD9 pKa = 3.75ALQEE13 pKa = 4.0NLLMLYY19 pKa = 9.85EE20 pKa = 4.8AGSDD24 pKa = 4.08KK25 pKa = 10.79IDD27 pKa = 3.73DD28 pKa = 4.01QIMYY32 pKa = 8.91WDD34 pKa = 3.98IMRR37 pKa = 11.84QEE39 pKa = 4.13NVLFYY44 pKa = 9.99YY45 pKa = 10.48ARR47 pKa = 11.84KK48 pKa = 10.04KK49 pKa = 10.43NIARR53 pKa = 11.84IGLQPVPSLTTSEE66 pKa = 4.88HH67 pKa = 6.17KK68 pKa = 10.68AKK70 pKa = 10.06QAIMMSLQLKK80 pKa = 7.54SLKK83 pKa = 9.52MSAYY87 pKa = 9.9GKK89 pKa = 10.05EE90 pKa = 3.89PWTLQDD96 pKa = 3.25TSFEE100 pKa = 4.48VYY102 pKa = 9.96NAAPFNTFKK111 pKa = 10.87KK112 pKa = 10.42GAFTVDD118 pKa = 2.58VFYY121 pKa = 11.15DD122 pKa = 3.89DD123 pKa = 6.53DD124 pKa = 4.21EE125 pKa = 6.68DD126 pKa = 3.97NYY128 pKa = 11.36YY129 pKa = 10.15PYY131 pKa = 9.83TAWSFIYY138 pKa = 10.56YY139 pKa = 10.32EE140 pKa = 4.35NGDD143 pKa = 3.95GRR145 pKa = 11.84WHH147 pKa = 6.17KK148 pKa = 11.25VEE150 pKa = 4.13GQADD154 pKa = 3.86YY155 pKa = 11.11EE156 pKa = 4.14GLYY159 pKa = 10.48FISHH163 pKa = 7.16DD164 pKa = 3.79NEE166 pKa = 3.49KK167 pKa = 10.79NYY169 pKa = 10.86YY170 pKa = 8.23VTFDD174 pKa = 3.04TDD176 pKa = 3.04APRR179 pKa = 11.84FSRR182 pKa = 11.84TGHH185 pKa = 4.67WTVKK189 pKa = 10.1YY190 pKa = 9.79KK191 pKa = 10.9NHH193 pKa = 6.95TISSTSVTSTSGNPAGDD210 pKa = 3.53PSSRR214 pKa = 11.84EE215 pKa = 3.85GTNTVGQEE223 pKa = 4.32TPVSQTNRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84QRR235 pKa = 11.84EE236 pKa = 4.2SSSSPNRR243 pKa = 11.84RR244 pKa = 11.84TRR246 pKa = 11.84QTTSQSDD253 pKa = 3.62WDD255 pKa = 4.3PDD257 pKa = 3.67SSPSSGEE264 pKa = 3.48RR265 pKa = 11.84LPRR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84EE274 pKa = 3.29RR275 pKa = 11.84APAAKK280 pKa = 9.72RR281 pKa = 11.84RR282 pKa = 11.84EE283 pKa = 4.07QRR285 pKa = 11.84KK286 pKa = 6.78STKK289 pKa = 10.42SLVPSPEE296 pKa = 3.95EE297 pKa = 3.35VGRR300 pKa = 11.84RR301 pKa = 11.84HH302 pKa = 6.36KK303 pKa = 10.82LVEE306 pKa = 4.26GKK308 pKa = 10.35GLSRR312 pKa = 11.84LRR314 pKa = 11.84RR315 pKa = 11.84LQEE318 pKa = 4.04EE319 pKa = 4.15AWDD322 pKa = 4.07PPVLLLKK329 pKa = 10.96GPANTLKK336 pKa = 10.55CYY338 pKa = 10.28RR339 pKa = 11.84YY340 pKa = 9.59RR341 pKa = 11.84CKK343 pKa = 10.23PRR345 pKa = 11.84CKK347 pKa = 10.51GLFTRR352 pKa = 11.84MSTVFSWVGEE362 pKa = 3.86GSEE365 pKa = 5.42RR366 pKa = 11.84IGQPRR371 pKa = 11.84MLLAFANNEE380 pKa = 3.84QRR382 pKa = 11.84RR383 pKa = 11.84HH384 pKa = 6.14FINTVHH390 pKa = 6.53LPKK393 pKa = 10.15GTEE396 pKa = 3.88YY397 pKa = 11.24SLGRR401 pKa = 11.84LDD403 pKa = 4.6SLL405 pKa = 4.66
MM1 pKa = 6.94EE2 pKa = 4.99TLNEE6 pKa = 4.24RR7 pKa = 11.84YY8 pKa = 9.3DD9 pKa = 3.75ALQEE13 pKa = 4.0NLLMLYY19 pKa = 9.85EE20 pKa = 4.8AGSDD24 pKa = 4.08KK25 pKa = 10.79IDD27 pKa = 3.73DD28 pKa = 4.01QIMYY32 pKa = 8.91WDD34 pKa = 3.98IMRR37 pKa = 11.84QEE39 pKa = 4.13NVLFYY44 pKa = 9.99YY45 pKa = 10.48ARR47 pKa = 11.84KK48 pKa = 10.04KK49 pKa = 10.43NIARR53 pKa = 11.84IGLQPVPSLTTSEE66 pKa = 4.88HH67 pKa = 6.17KK68 pKa = 10.68AKK70 pKa = 10.06QAIMMSLQLKK80 pKa = 7.54SLKK83 pKa = 9.52MSAYY87 pKa = 9.9GKK89 pKa = 10.05EE90 pKa = 3.89PWTLQDD96 pKa = 3.25TSFEE100 pKa = 4.48VYY102 pKa = 9.96NAAPFNTFKK111 pKa = 10.87KK112 pKa = 10.42GAFTVDD118 pKa = 2.58VFYY121 pKa = 11.15DD122 pKa = 3.89DD123 pKa = 6.53DD124 pKa = 4.21EE125 pKa = 6.68DD126 pKa = 3.97NYY128 pKa = 11.36YY129 pKa = 10.15PYY131 pKa = 9.83TAWSFIYY138 pKa = 10.56YY139 pKa = 10.32EE140 pKa = 4.35NGDD143 pKa = 3.95GRR145 pKa = 11.84WHH147 pKa = 6.17KK148 pKa = 11.25VEE150 pKa = 4.13GQADD154 pKa = 3.86YY155 pKa = 11.11EE156 pKa = 4.14GLYY159 pKa = 10.48FISHH163 pKa = 7.16DD164 pKa = 3.79NEE166 pKa = 3.49KK167 pKa = 10.79NYY169 pKa = 10.86YY170 pKa = 8.23VTFDD174 pKa = 3.04TDD176 pKa = 3.04APRR179 pKa = 11.84FSRR182 pKa = 11.84TGHH185 pKa = 4.67WTVKK189 pKa = 10.1YY190 pKa = 9.79KK191 pKa = 10.9NHH193 pKa = 6.95TISSTSVTSTSGNPAGDD210 pKa = 3.53PSSRR214 pKa = 11.84EE215 pKa = 3.85GTNTVGQEE223 pKa = 4.32TPVSQTNRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84QRR235 pKa = 11.84EE236 pKa = 4.2SSSSPNRR243 pKa = 11.84RR244 pKa = 11.84TRR246 pKa = 11.84QTTSQSDD253 pKa = 3.62WDD255 pKa = 4.3PDD257 pKa = 3.67SSPSSGEE264 pKa = 3.48RR265 pKa = 11.84LPRR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84EE274 pKa = 3.29RR275 pKa = 11.84APAAKK280 pKa = 9.72RR281 pKa = 11.84RR282 pKa = 11.84EE283 pKa = 4.07QRR285 pKa = 11.84KK286 pKa = 6.78STKK289 pKa = 10.42SLVPSPEE296 pKa = 3.95EE297 pKa = 3.35VGRR300 pKa = 11.84RR301 pKa = 11.84HH302 pKa = 6.36KK303 pKa = 10.82LVEE306 pKa = 4.26GKK308 pKa = 10.35GLSRR312 pKa = 11.84LRR314 pKa = 11.84RR315 pKa = 11.84LQEE318 pKa = 4.04EE319 pKa = 4.15AWDD322 pKa = 4.07PPVLLLKK329 pKa = 10.96GPANTLKK336 pKa = 10.55CYY338 pKa = 10.28RR339 pKa = 11.84YY340 pKa = 9.59RR341 pKa = 11.84CKK343 pKa = 10.23PRR345 pKa = 11.84CKK347 pKa = 10.51GLFTRR352 pKa = 11.84MSTVFSWVGEE362 pKa = 3.86GSEE365 pKa = 5.42RR366 pKa = 11.84IGQPRR371 pKa = 11.84MLLAFANNEE380 pKa = 3.84QRR382 pKa = 11.84RR383 pKa = 11.84HH384 pKa = 6.14FINTVHH390 pKa = 6.53LPKK393 pKa = 10.15GTEE396 pKa = 3.88YY397 pKa = 11.24SLGRR401 pKa = 11.84LDD403 pKa = 4.6SLL405 pKa = 4.66
Molecular weight: 46.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2426 |
93 |
617 |
346.6 |
38.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.73 ± 0.414 | 2.102 ± 0.754 |
6.595 ± 0.404 | 6.183 ± 0.692 |
4.534 ± 0.478 | 5.771 ± 0.621 |
1.855 ± 0.183 | 4.864 ± 0.571 |
5.111 ± 0.94 | 8.862 ± 0.677 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.649 ± 0.316 | 5.276 ± 0.38 |
5.894 ± 0.954 | 4.122 ± 0.55 |
5.936 ± 1.161 | 8.45 ± 0.509 |
6.183 ± 0.614 | 6.636 ± 0.721 |
1.237 ± 0.279 | 3.009 ± 0.5 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
