
Beet western yellows virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 8.3
Get precalculated fractions of proteins
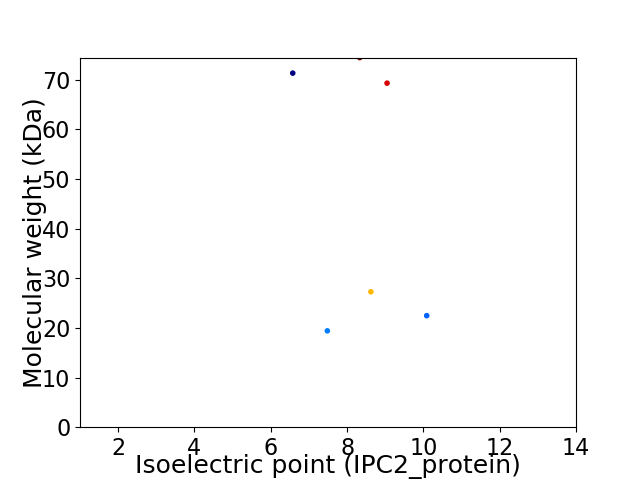
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q8JZ11|Q8JZ11_9LUTE Serine protease OS=Beet western yellows virus OX=12042 PE=4 SV=1
SS1 pKa = 7.6GDD3 pKa = 3.45RR4 pKa = 11.84QEE6 pKa = 4.12LGKK9 pKa = 10.38RR10 pKa = 11.84EE11 pKa = 4.15SGSARR16 pKa = 11.84RR17 pKa = 11.84NNRR20 pKa = 11.84NPQRR24 pKa = 11.84KK25 pKa = 8.26HH26 pKa = 4.39PQGRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 7.95QWRR35 pKa = 11.84RR36 pKa = 11.84NPPSCCSYY44 pKa = 9.59FTEE47 pKa = 4.74GTLGPRR53 pKa = 11.84CHH55 pKa = 6.92ASHH58 pKa = 6.26VHH60 pKa = 5.52TPDD63 pKa = 5.51DD64 pKa = 4.38EE65 pKa = 4.55GQPGCWCEE73 pKa = 4.4VPNHH77 pKa = 6.67DD78 pKa = 4.18CFFRR82 pKa = 11.84RR83 pKa = 11.84FQRR86 pKa = 11.84DD87 pKa = 3.52QRR89 pKa = 11.84SHH91 pKa = 6.42PAEE94 pKa = 3.99DD95 pKa = 4.05RR96 pKa = 11.84YY97 pKa = 11.55SFDD100 pKa = 3.64RR101 pKa = 11.84EE102 pKa = 4.0AGRR105 pKa = 11.84GDD107 pKa = 3.38PCRR110 pKa = 11.84EE111 pKa = 4.18GNEE114 pKa = 3.96EE115 pKa = 4.17TPEE118 pKa = 3.84SRR120 pKa = 11.84AEE122 pKa = 4.24KK123 pKa = 9.93ICEE126 pKa = 3.73QAEE129 pKa = 4.21NFCQYY134 pKa = 10.52FSSLYY139 pKa = 9.16RR140 pKa = 11.84WEE142 pKa = 4.41SGSGKK147 pKa = 9.24EE148 pKa = 4.47APGFEE153 pKa = 4.62QIGSLPQFYY162 pKa = 10.3HH163 pKa = 6.83PKK165 pKa = 10.02QKK167 pKa = 11.06GEE169 pKa = 4.25SRR171 pKa = 11.84WGAKK175 pKa = 8.9VLQLHH180 pKa = 7.05PEE182 pKa = 4.34LEE184 pKa = 4.15AQTRR188 pKa = 11.84GFGWPQFGAQAEE200 pKa = 4.48LKK202 pKa = 10.45SLRR205 pKa = 11.84LQAARR210 pKa = 11.84WLEE213 pKa = 3.69RR214 pKa = 11.84AQQVKK219 pKa = 10.17IPSTEE224 pKa = 3.39EE225 pKa = 3.52RR226 pKa = 11.84EE227 pKa = 3.96RR228 pKa = 11.84VIRR231 pKa = 11.84KK232 pKa = 9.1CCEE235 pKa = 3.37AYY237 pKa = 10.63QNAKK241 pKa = 10.68SNGPNATRR249 pKa = 11.84GNKK252 pKa = 9.79LSWDD256 pKa = 3.5NFLEE260 pKa = 4.26DD261 pKa = 4.1FKK263 pKa = 11.41QAVFSLEE270 pKa = 3.42FDD272 pKa = 3.34AGIGVPYY279 pKa = 9.92IAYY282 pKa = 8.45GKK284 pKa = 6.46PTHH287 pKa = 6.61RR288 pKa = 11.84GWVEE292 pKa = 3.85DD293 pKa = 3.95QEE295 pKa = 4.56LLPILARR302 pKa = 11.84LTFIRR307 pKa = 11.84LQKK310 pKa = 9.52MLEE313 pKa = 3.84ARR315 pKa = 11.84FEE317 pKa = 4.18EE318 pKa = 4.71LSPEE322 pKa = 4.01QLVQEE327 pKa = 4.7GLCDD331 pKa = 4.87PIRR334 pKa = 11.84VFVKK338 pKa = 10.78GEE340 pKa = 3.59PHH342 pKa = 6.19KK343 pKa = 10.54QSKK346 pKa = 10.21LDD348 pKa = 3.38EE349 pKa = 4.27GRR351 pKa = 11.84YY352 pKa = 8.96RR353 pKa = 11.84LIMSVSLVDD362 pKa = 3.45QLVARR367 pKa = 11.84VLFQNQNKK375 pKa = 9.66RR376 pKa = 11.84EE377 pKa = 3.95IALWRR382 pKa = 11.84AVPSKK387 pKa = 10.57PGFGLSTDD395 pKa = 3.6EE396 pKa = 3.99QVVEE400 pKa = 4.75FVQMLSAQVKK410 pKa = 8.85VRR412 pKa = 11.84PPQLISDD419 pKa = 4.26WRR421 pKa = 11.84QHH423 pKa = 6.52LVATDD428 pKa = 3.61CSGFDD433 pKa = 3.32WSVSDD438 pKa = 4.58WLLEE442 pKa = 4.07DD443 pKa = 4.28DD444 pKa = 3.82MEE446 pKa = 4.4VRR448 pKa = 11.84NRR450 pKa = 11.84LTTDD454 pKa = 3.34LNEE457 pKa = 4.31TTSKK461 pKa = 10.7LRR463 pKa = 11.84AAWLKK468 pKa = 10.69CISNSVLCLSDD479 pKa = 3.31GTLLAQRR486 pKa = 11.84VPGVQKK492 pKa = 10.45SGSYY496 pKa = 8.25NTSSSNSRR504 pKa = 11.84IRR506 pKa = 11.84VMAAFHH512 pKa = 6.86CGAEE516 pKa = 3.94WAMAMGDD523 pKa = 3.77DD524 pKa = 4.0ALEE527 pKa = 4.35SVSTDD532 pKa = 3.08LGKK535 pKa = 10.78YY536 pKa = 8.84AALGFKK542 pKa = 10.44VEE544 pKa = 4.31EE545 pKa = 4.12SSKK548 pKa = 11.47LEE550 pKa = 3.98FCSHH554 pKa = 5.14IFEE557 pKa = 5.18RR558 pKa = 11.84EE559 pKa = 3.87DD560 pKa = 3.57LAVPVNKK567 pKa = 10.6AKK569 pKa = 10.09MIYY572 pKa = 10.08KK573 pKa = 9.93LIHH576 pKa = 6.3GYY578 pKa = 9.39EE579 pKa = 4.21PEE581 pKa = 4.62CGNPEE586 pKa = 4.01VLVNYY591 pKa = 7.63LTACFAILNEE601 pKa = 4.31LRR603 pKa = 11.84SDD605 pKa = 3.53PQMVQTLYY613 pKa = 10.34SWLVKK618 pKa = 9.87PVQPQKK624 pKa = 11.28NN625 pKa = 3.49
SS1 pKa = 7.6GDD3 pKa = 3.45RR4 pKa = 11.84QEE6 pKa = 4.12LGKK9 pKa = 10.38RR10 pKa = 11.84EE11 pKa = 4.15SGSARR16 pKa = 11.84RR17 pKa = 11.84NNRR20 pKa = 11.84NPQRR24 pKa = 11.84KK25 pKa = 8.26HH26 pKa = 4.39PQGRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 7.95QWRR35 pKa = 11.84RR36 pKa = 11.84NPPSCCSYY44 pKa = 9.59FTEE47 pKa = 4.74GTLGPRR53 pKa = 11.84CHH55 pKa = 6.92ASHH58 pKa = 6.26VHH60 pKa = 5.52TPDD63 pKa = 5.51DD64 pKa = 4.38EE65 pKa = 4.55GQPGCWCEE73 pKa = 4.4VPNHH77 pKa = 6.67DD78 pKa = 4.18CFFRR82 pKa = 11.84RR83 pKa = 11.84FQRR86 pKa = 11.84DD87 pKa = 3.52QRR89 pKa = 11.84SHH91 pKa = 6.42PAEE94 pKa = 3.99DD95 pKa = 4.05RR96 pKa = 11.84YY97 pKa = 11.55SFDD100 pKa = 3.64RR101 pKa = 11.84EE102 pKa = 4.0AGRR105 pKa = 11.84GDD107 pKa = 3.38PCRR110 pKa = 11.84EE111 pKa = 4.18GNEE114 pKa = 3.96EE115 pKa = 4.17TPEE118 pKa = 3.84SRR120 pKa = 11.84AEE122 pKa = 4.24KK123 pKa = 9.93ICEE126 pKa = 3.73QAEE129 pKa = 4.21NFCQYY134 pKa = 10.52FSSLYY139 pKa = 9.16RR140 pKa = 11.84WEE142 pKa = 4.41SGSGKK147 pKa = 9.24EE148 pKa = 4.47APGFEE153 pKa = 4.62QIGSLPQFYY162 pKa = 10.3HH163 pKa = 6.83PKK165 pKa = 10.02QKK167 pKa = 11.06GEE169 pKa = 4.25SRR171 pKa = 11.84WGAKK175 pKa = 8.9VLQLHH180 pKa = 7.05PEE182 pKa = 4.34LEE184 pKa = 4.15AQTRR188 pKa = 11.84GFGWPQFGAQAEE200 pKa = 4.48LKK202 pKa = 10.45SLRR205 pKa = 11.84LQAARR210 pKa = 11.84WLEE213 pKa = 3.69RR214 pKa = 11.84AQQVKK219 pKa = 10.17IPSTEE224 pKa = 3.39EE225 pKa = 3.52RR226 pKa = 11.84EE227 pKa = 3.96RR228 pKa = 11.84VIRR231 pKa = 11.84KK232 pKa = 9.1CCEE235 pKa = 3.37AYY237 pKa = 10.63QNAKK241 pKa = 10.68SNGPNATRR249 pKa = 11.84GNKK252 pKa = 9.79LSWDD256 pKa = 3.5NFLEE260 pKa = 4.26DD261 pKa = 4.1FKK263 pKa = 11.41QAVFSLEE270 pKa = 3.42FDD272 pKa = 3.34AGIGVPYY279 pKa = 9.92IAYY282 pKa = 8.45GKK284 pKa = 6.46PTHH287 pKa = 6.61RR288 pKa = 11.84GWVEE292 pKa = 3.85DD293 pKa = 3.95QEE295 pKa = 4.56LLPILARR302 pKa = 11.84LTFIRR307 pKa = 11.84LQKK310 pKa = 9.52MLEE313 pKa = 3.84ARR315 pKa = 11.84FEE317 pKa = 4.18EE318 pKa = 4.71LSPEE322 pKa = 4.01QLVQEE327 pKa = 4.7GLCDD331 pKa = 4.87PIRR334 pKa = 11.84VFVKK338 pKa = 10.78GEE340 pKa = 3.59PHH342 pKa = 6.19KK343 pKa = 10.54QSKK346 pKa = 10.21LDD348 pKa = 3.38EE349 pKa = 4.27GRR351 pKa = 11.84YY352 pKa = 8.96RR353 pKa = 11.84LIMSVSLVDD362 pKa = 3.45QLVARR367 pKa = 11.84VLFQNQNKK375 pKa = 9.66RR376 pKa = 11.84EE377 pKa = 3.95IALWRR382 pKa = 11.84AVPSKK387 pKa = 10.57PGFGLSTDD395 pKa = 3.6EE396 pKa = 3.99QVVEE400 pKa = 4.75FVQMLSAQVKK410 pKa = 8.85VRR412 pKa = 11.84PPQLISDD419 pKa = 4.26WRR421 pKa = 11.84QHH423 pKa = 6.52LVATDD428 pKa = 3.61CSGFDD433 pKa = 3.32WSVSDD438 pKa = 4.58WLLEE442 pKa = 4.07DD443 pKa = 4.28DD444 pKa = 3.82MEE446 pKa = 4.4VRR448 pKa = 11.84NRR450 pKa = 11.84LTTDD454 pKa = 3.34LNEE457 pKa = 4.31TTSKK461 pKa = 10.7LRR463 pKa = 11.84AAWLKK468 pKa = 10.69CISNSVLCLSDD479 pKa = 3.31GTLLAQRR486 pKa = 11.84VPGVQKK492 pKa = 10.45SGSYY496 pKa = 8.25NTSSSNSRR504 pKa = 11.84IRR506 pKa = 11.84VMAAFHH512 pKa = 6.86CGAEE516 pKa = 3.94WAMAMGDD523 pKa = 3.77DD524 pKa = 4.0ALEE527 pKa = 4.35SVSTDD532 pKa = 3.08LGKK535 pKa = 10.78YY536 pKa = 8.84AALGFKK542 pKa = 10.44VEE544 pKa = 4.31EE545 pKa = 4.12SSKK548 pKa = 11.47LEE550 pKa = 3.98FCSHH554 pKa = 5.14IFEE557 pKa = 5.18RR558 pKa = 11.84EE559 pKa = 3.87DD560 pKa = 3.57LAVPVNKK567 pKa = 10.6AKK569 pKa = 10.09MIYY572 pKa = 10.08KK573 pKa = 9.93LIHH576 pKa = 6.3GYY578 pKa = 9.39EE579 pKa = 4.21PEE581 pKa = 4.62CGNPEE586 pKa = 4.01VLVNYY591 pKa = 7.63LTACFAILNEE601 pKa = 4.31LRR603 pKa = 11.84SDD605 pKa = 3.53PQMVQTLYY613 pKa = 10.34SWLVKK618 pKa = 9.87PVQPQKK624 pKa = 11.28NN625 pKa = 3.49
Molecular weight: 71.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8JZ09|Q8JZ09_9LUTE Readthrough protein OS=Beet western yellows virus OX=12042 PE=3 SV=1
MM1 pKa = 6.49NTVVGRR7 pKa = 11.84RR8 pKa = 11.84TINGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84QARR21 pKa = 11.84RR22 pKa = 11.84AQRR25 pKa = 11.84SQPVVVVQTSRR36 pKa = 11.84TTQRR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GNNRR51 pKa = 11.84TRR53 pKa = 11.84GAVSTRR59 pKa = 11.84GASSSEE65 pKa = 3.81TFVFSKK71 pKa = 11.13DD72 pKa = 3.3NLAGSSSGAITFGPSLSDD90 pKa = 4.1CPAFSNGILKK100 pKa = 10.26AYY102 pKa = 10.04HH103 pKa = 6.58EE104 pKa = 4.78YY105 pKa = 10.01KK106 pKa = 10.07ISMVILEE113 pKa = 4.63FVSEE117 pKa = 4.2ASSQNSGSIAYY128 pKa = 9.28EE129 pKa = 3.87LDD131 pKa = 3.1PHH133 pKa = 6.75CRR135 pKa = 11.84LDD137 pKa = 4.04ALSSTINKK145 pKa = 9.79FGITKK150 pKa = 9.13PGRR153 pKa = 11.84RR154 pKa = 11.84AFTASYY160 pKa = 10.99INGTDD165 pKa = 2.79WHH167 pKa = 7.19DD168 pKa = 3.51VAKK171 pKa = 10.53DD172 pKa = 3.32QFRR175 pKa = 11.84ILYY178 pKa = 8.84KK179 pKa = 11.0GNGSSSIAGSFRR191 pKa = 11.84ITMKK195 pKa = 10.75CQFHH199 pKa = 6.24NPKK202 pKa = 10.51
MM1 pKa = 6.49NTVVGRR7 pKa = 11.84RR8 pKa = 11.84TINGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84QARR21 pKa = 11.84RR22 pKa = 11.84AQRR25 pKa = 11.84SQPVVVVQTSRR36 pKa = 11.84TTQRR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GNNRR51 pKa = 11.84TRR53 pKa = 11.84GAVSTRR59 pKa = 11.84GASSSEE65 pKa = 3.81TFVFSKK71 pKa = 11.13DD72 pKa = 3.3NLAGSSSGAITFGPSLSDD90 pKa = 4.1CPAFSNGILKK100 pKa = 10.26AYY102 pKa = 10.04HH103 pKa = 6.58EE104 pKa = 4.78YY105 pKa = 10.01KK106 pKa = 10.07ISMVILEE113 pKa = 4.63FVSEE117 pKa = 4.2ASSQNSGSIAYY128 pKa = 9.28EE129 pKa = 3.87LDD131 pKa = 3.1PHH133 pKa = 6.75CRR135 pKa = 11.84LDD137 pKa = 4.04ALSSTINKK145 pKa = 9.79FGITKK150 pKa = 9.13PGRR153 pKa = 11.84RR154 pKa = 11.84AFTASYY160 pKa = 10.99INGTDD165 pKa = 2.79WHH167 pKa = 7.19DD168 pKa = 3.51VAKK171 pKa = 10.53DD172 pKa = 3.32QFRR175 pKa = 11.84ILYY178 pKa = 8.84KK179 pKa = 11.0GNGSSSIAGSFRR191 pKa = 11.84ITMKK195 pKa = 10.75CQFHH199 pKa = 6.24NPKK202 pKa = 10.51
Molecular weight: 22.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2545 |
175 |
670 |
424.2 |
47.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.269 ± 0.526 | 1.768 ± 0.412 |
4.086 ± 0.788 | 6.09 ± 0.762 |
3.929 ± 0.37 | 6.798 ± 0.242 |
2.24 ± 0.329 | 4.126 ± 0.479 |
5.383 ± 0.454 | 7.583 ± 1.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.965 ± 0.234 | 4.283 ± 0.437 |
5.815 ± 0.514 | 4.715 ± 0.575 |
7.623 ± 0.887 | 9.98 ± 0.933 |
6.051 ± 1.051 | 5.855 ± 0.334 |
1.768 ± 0.234 | 2.633 ± 0.18 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
