
Aspergillus mulundensis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina;
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
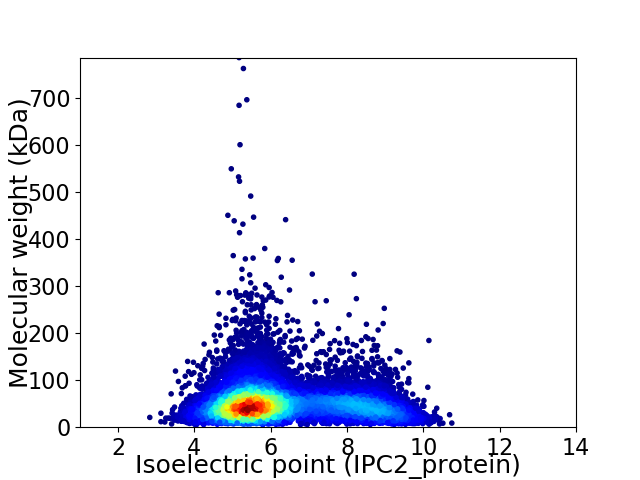
Virtual 2D-PAGE plot for 11569 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3D8R417|A0A3D8R417_9EURO Cupin_2 domain-containing protein OS=Aspergillus mulundensis OX=1810919 GN=DSM5745_08562 PE=4 SV=1
MM1 pKa = 7.31YY2 pKa = 9.98YY3 pKa = 11.09ALGSLALVAFATEE16 pKa = 4.54VLATPHH22 pKa = 6.18PMITASPTLAKK33 pKa = 9.93RR34 pKa = 11.84DD35 pKa = 3.41SCTFSGSDD43 pKa = 3.46GAASASKK50 pKa = 10.4SQADD54 pKa = 4.03CATITLSDD62 pKa = 3.27ITVPSGTTLDD72 pKa = 4.41LSDD75 pKa = 5.7LEE77 pKa = 6.28DD78 pKa = 3.8DD79 pKa = 3.79TTVVFEE85 pKa = 4.8GTTSWEE91 pKa = 4.01YY92 pKa = 11.19EE93 pKa = 4.1EE94 pKa = 4.73WDD96 pKa = 4.42GPLLQIKK103 pKa = 8.39GTGITIKK110 pKa = 10.69GADD113 pKa = 3.79GAKK116 pKa = 10.27LNPDD120 pKa = 3.7GSRR123 pKa = 11.84WWDD126 pKa = 3.4GEE128 pKa = 3.98GSNGGVTKK136 pKa = 10.54PKK138 pKa = 10.33FFYY141 pKa = 10.56AHH143 pKa = 6.09SLKK146 pKa = 10.95DD147 pKa = 3.43STIEE151 pKa = 3.61NLYY154 pKa = 10.36IEE156 pKa = 4.64NTPVQAVSINSCDD169 pKa = 3.44GLTITDD175 pKa = 3.54MTIDD179 pKa = 3.6NSAGDD184 pKa = 3.91DD185 pKa = 3.66AGGHH189 pKa = 5.07NTDD192 pKa = 4.03GFDD195 pKa = 3.12IGEE198 pKa = 4.42SNNVVITGAKK208 pKa = 9.79VYY210 pKa = 10.84NQDD213 pKa = 3.04DD214 pKa = 4.2CVAVNSGTEE223 pKa = 3.75ITFSGGTCSGGHH235 pKa = 5.98GLSIGSVGGRR245 pKa = 11.84DD246 pKa = 4.03DD247 pKa = 3.81NTVDD251 pKa = 3.1TVTFKK256 pKa = 11.29DD257 pKa = 3.66STVSNSVNGIRR268 pKa = 11.84IKK270 pKa = 10.87AKK272 pKa = 9.98SDD274 pKa = 3.02EE275 pKa = 4.2TGEE278 pKa = 4.34IKK280 pKa = 10.66GVTYY284 pKa = 10.78SGITLDD290 pKa = 4.48SISKK294 pKa = 9.82YY295 pKa = 10.83GILIEE300 pKa = 3.98QNYY303 pKa = 10.68DD304 pKa = 3.16GGDD307 pKa = 3.43LHH309 pKa = 7.46GDD311 pKa = 3.57PTSGIPITDD320 pKa = 3.6LTIEE324 pKa = 5.08DD325 pKa = 3.77ISGSGAVDD333 pKa = 3.11SDD335 pKa = 3.93GYY337 pKa = 11.22NIVVVCGDD345 pKa = 4.74DD346 pKa = 4.43GCSDD350 pKa = 3.61WTWSGVEE357 pKa = 3.83VSGGEE362 pKa = 4.52DD363 pKa = 3.46YY364 pKa = 11.38SDD366 pKa = 4.34CEE368 pKa = 4.3NVPSVASCSTT378 pKa = 3.26
MM1 pKa = 7.31YY2 pKa = 9.98YY3 pKa = 11.09ALGSLALVAFATEE16 pKa = 4.54VLATPHH22 pKa = 6.18PMITASPTLAKK33 pKa = 9.93RR34 pKa = 11.84DD35 pKa = 3.41SCTFSGSDD43 pKa = 3.46GAASASKK50 pKa = 10.4SQADD54 pKa = 4.03CATITLSDD62 pKa = 3.27ITVPSGTTLDD72 pKa = 4.41LSDD75 pKa = 5.7LEE77 pKa = 6.28DD78 pKa = 3.8DD79 pKa = 3.79TTVVFEE85 pKa = 4.8GTTSWEE91 pKa = 4.01YY92 pKa = 11.19EE93 pKa = 4.1EE94 pKa = 4.73WDD96 pKa = 4.42GPLLQIKK103 pKa = 8.39GTGITIKK110 pKa = 10.69GADD113 pKa = 3.79GAKK116 pKa = 10.27LNPDD120 pKa = 3.7GSRR123 pKa = 11.84WWDD126 pKa = 3.4GEE128 pKa = 3.98GSNGGVTKK136 pKa = 10.54PKK138 pKa = 10.33FFYY141 pKa = 10.56AHH143 pKa = 6.09SLKK146 pKa = 10.95DD147 pKa = 3.43STIEE151 pKa = 3.61NLYY154 pKa = 10.36IEE156 pKa = 4.64NTPVQAVSINSCDD169 pKa = 3.44GLTITDD175 pKa = 3.54MTIDD179 pKa = 3.6NSAGDD184 pKa = 3.91DD185 pKa = 3.66AGGHH189 pKa = 5.07NTDD192 pKa = 4.03GFDD195 pKa = 3.12IGEE198 pKa = 4.42SNNVVITGAKK208 pKa = 9.79VYY210 pKa = 10.84NQDD213 pKa = 3.04DD214 pKa = 4.2CVAVNSGTEE223 pKa = 3.75ITFSGGTCSGGHH235 pKa = 5.98GLSIGSVGGRR245 pKa = 11.84DD246 pKa = 4.03DD247 pKa = 3.81NTVDD251 pKa = 3.1TVTFKK256 pKa = 11.29DD257 pKa = 3.66STVSNSVNGIRR268 pKa = 11.84IKK270 pKa = 10.87AKK272 pKa = 9.98SDD274 pKa = 3.02EE275 pKa = 4.2TGEE278 pKa = 4.34IKK280 pKa = 10.66GVTYY284 pKa = 10.78SGITLDD290 pKa = 4.48SISKK294 pKa = 9.82YY295 pKa = 10.83GILIEE300 pKa = 3.98QNYY303 pKa = 10.68DD304 pKa = 3.16GGDD307 pKa = 3.43LHH309 pKa = 7.46GDD311 pKa = 3.57PTSGIPITDD320 pKa = 3.6LTIEE324 pKa = 5.08DD325 pKa = 3.77ISGSGAVDD333 pKa = 3.11SDD335 pKa = 3.93GYY337 pKa = 11.22NIVVVCGDD345 pKa = 4.74DD346 pKa = 4.43GCSDD350 pKa = 3.61WTWSGVEE357 pKa = 3.83VSGGEE362 pKa = 4.52DD363 pKa = 3.46YY364 pKa = 11.38SDD366 pKa = 4.34CEE368 pKa = 4.3NVPSVASCSTT378 pKa = 3.26
Molecular weight: 39.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3D8QJG5|A0A3D8QJG5_9EURO Ankyrin repeat protein OS=Aspergillus mulundensis OX=1810919 GN=DSM5745_10492 PE=4 SV=1
MM1 pKa = 7.68SIGAYY6 pKa = 8.58EE7 pKa = 4.02VVMCGEE13 pKa = 4.77DD14 pKa = 3.6EE15 pKa = 4.07QSEE18 pKa = 4.28ALISPSHH25 pKa = 6.61RR26 pKa = 11.84NGFKK30 pKa = 10.05PAPMCKK36 pKa = 9.44DD37 pKa = 3.72DD38 pKa = 3.61NHH40 pKa = 6.71GATEE44 pKa = 4.02YY45 pKa = 10.03WISEE49 pKa = 4.27SEE51 pKa = 4.27RR52 pKa = 11.84PHH54 pKa = 5.8PRR56 pKa = 11.84AVRR59 pKa = 11.84SSTNEE64 pKa = 3.54PAHH67 pKa = 6.51PRR69 pKa = 11.84TPSPPKK75 pKa = 10.36HH76 pKa = 6.22RR77 pKa = 11.84VTTCSQTVRR86 pKa = 11.84RR87 pKa = 11.84CQTHH91 pKa = 6.27IGIHH95 pKa = 5.33HH96 pKa = 7.22RR97 pKa = 11.84NNKK100 pKa = 8.28GAPAHH105 pKa = 5.76QNPDD109 pKa = 3.03VAPRR113 pKa = 11.84KK114 pKa = 9.31ARR116 pKa = 11.84HH117 pKa = 5.93APFPRR122 pKa = 11.84PPHH125 pKa = 5.63PLRR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84LQCIAHH136 pKa = 6.51AFGYY140 pKa = 8.24TPHH143 pKa = 7.21EE144 pKa = 4.57IIGIVFDD151 pKa = 4.83RR152 pKa = 11.84PVLAPNMAPANEE164 pKa = 4.04ALKK167 pKa = 10.2EE168 pKa = 3.88ARR170 pKa = 11.84IYY172 pKa = 10.36NHH174 pKa = 6.35NEE176 pKa = 3.62SPRR179 pKa = 11.84PARR182 pKa = 11.84AGVYY186 pKa = 10.48LLLLHH191 pKa = 7.13RR192 pKa = 11.84APATGGEE199 pKa = 4.27TTIPSSLEE207 pKa = 3.41LFRR210 pKa = 11.84RR211 pKa = 11.84ARR213 pKa = 11.84AEE215 pKa = 3.93MPTSLTCWRR224 pKa = 11.84RR225 pKa = 11.84GHH227 pKa = 6.6HH228 pKa = 6.22EE229 pKa = 4.19PGHH232 pKa = 5.45VYY234 pKa = 10.27GRR236 pKa = 11.84AAVRR240 pKa = 11.84GRR242 pKa = 11.84VDD244 pKa = 4.02DD245 pKa = 3.72EE246 pKa = 4.3TGVRR250 pKa = 11.84KK251 pKa = 9.77EE252 pKa = 4.55FADD255 pKa = 3.74TDD257 pKa = 3.77DD258 pKa = 6.06DD259 pKa = 3.7EE260 pKa = 4.69TKK262 pKa = 10.25RR263 pKa = 11.84RR264 pKa = 11.84KK265 pKa = 9.91VEE267 pKa = 3.82AQIARR272 pKa = 11.84YY273 pKa = 9.53GRR275 pKa = 11.84GRR277 pKa = 11.84HH278 pKa = 4.95TTSEE282 pKa = 3.82WTDD285 pKa = 3.35DD286 pKa = 3.55DD287 pKa = 4.64RR288 pKa = 11.84EE289 pKa = 4.02RR290 pKa = 11.84LQLVLTHH297 pKa = 6.6HH298 pKa = 7.23LPRR301 pKa = 11.84VPSARR306 pKa = 11.84NPEE309 pKa = 4.05RR310 pKa = 11.84TFPLPTPFRR319 pKa = 11.84ASGVPQEE326 pKa = 4.53RR327 pKa = 11.84DD328 pKa = 2.78RR329 pKa = 11.84SQEE332 pKa = 3.89GKK334 pKa = 9.83SRR336 pKa = 11.84RR337 pKa = 11.84APGQEE342 pKa = 3.32RR343 pKa = 11.84RR344 pKa = 11.84ASGVWRR350 pKa = 11.84RR351 pKa = 11.84HH352 pKa = 5.2ADD354 pKa = 3.16SGGGPGGAGAHH365 pKa = 6.71HH366 pKa = 6.53GRR368 pKa = 11.84DD369 pKa = 3.52PRR371 pKa = 11.84ATSMAGGRR379 pKa = 11.84RR380 pKa = 11.84AGLRR384 pKa = 11.84QCRR387 pKa = 11.84CAAWAGAVGRR397 pKa = 11.84GAGGSRR403 pKa = 11.84CSGG406 pKa = 3.18
MM1 pKa = 7.68SIGAYY6 pKa = 8.58EE7 pKa = 4.02VVMCGEE13 pKa = 4.77DD14 pKa = 3.6EE15 pKa = 4.07QSEE18 pKa = 4.28ALISPSHH25 pKa = 6.61RR26 pKa = 11.84NGFKK30 pKa = 10.05PAPMCKK36 pKa = 9.44DD37 pKa = 3.72DD38 pKa = 3.61NHH40 pKa = 6.71GATEE44 pKa = 4.02YY45 pKa = 10.03WISEE49 pKa = 4.27SEE51 pKa = 4.27RR52 pKa = 11.84PHH54 pKa = 5.8PRR56 pKa = 11.84AVRR59 pKa = 11.84SSTNEE64 pKa = 3.54PAHH67 pKa = 6.51PRR69 pKa = 11.84TPSPPKK75 pKa = 10.36HH76 pKa = 6.22RR77 pKa = 11.84VTTCSQTVRR86 pKa = 11.84RR87 pKa = 11.84CQTHH91 pKa = 6.27IGIHH95 pKa = 5.33HH96 pKa = 7.22RR97 pKa = 11.84NNKK100 pKa = 8.28GAPAHH105 pKa = 5.76QNPDD109 pKa = 3.03VAPRR113 pKa = 11.84KK114 pKa = 9.31ARR116 pKa = 11.84HH117 pKa = 5.93APFPRR122 pKa = 11.84PPHH125 pKa = 5.63PLRR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84LQCIAHH136 pKa = 6.51AFGYY140 pKa = 8.24TPHH143 pKa = 7.21EE144 pKa = 4.57IIGIVFDD151 pKa = 4.83RR152 pKa = 11.84PVLAPNMAPANEE164 pKa = 4.04ALKK167 pKa = 10.2EE168 pKa = 3.88ARR170 pKa = 11.84IYY172 pKa = 10.36NHH174 pKa = 6.35NEE176 pKa = 3.62SPRR179 pKa = 11.84PARR182 pKa = 11.84AGVYY186 pKa = 10.48LLLLHH191 pKa = 7.13RR192 pKa = 11.84APATGGEE199 pKa = 4.27TTIPSSLEE207 pKa = 3.41LFRR210 pKa = 11.84RR211 pKa = 11.84ARR213 pKa = 11.84AEE215 pKa = 3.93MPTSLTCWRR224 pKa = 11.84RR225 pKa = 11.84GHH227 pKa = 6.6HH228 pKa = 6.22EE229 pKa = 4.19PGHH232 pKa = 5.45VYY234 pKa = 10.27GRR236 pKa = 11.84AAVRR240 pKa = 11.84GRR242 pKa = 11.84VDD244 pKa = 4.02DD245 pKa = 3.72EE246 pKa = 4.3TGVRR250 pKa = 11.84KK251 pKa = 9.77EE252 pKa = 4.55FADD255 pKa = 3.74TDD257 pKa = 3.77DD258 pKa = 6.06DD259 pKa = 3.7EE260 pKa = 4.69TKK262 pKa = 10.25RR263 pKa = 11.84RR264 pKa = 11.84KK265 pKa = 9.91VEE267 pKa = 3.82AQIARR272 pKa = 11.84YY273 pKa = 9.53GRR275 pKa = 11.84GRR277 pKa = 11.84HH278 pKa = 4.95TTSEE282 pKa = 3.82WTDD285 pKa = 3.35DD286 pKa = 3.55DD287 pKa = 4.64RR288 pKa = 11.84EE289 pKa = 4.02RR290 pKa = 11.84LQLVLTHH297 pKa = 6.6HH298 pKa = 7.23LPRR301 pKa = 11.84VPSARR306 pKa = 11.84NPEE309 pKa = 4.05RR310 pKa = 11.84TFPLPTPFRR319 pKa = 11.84ASGVPQEE326 pKa = 4.53RR327 pKa = 11.84DD328 pKa = 2.78RR329 pKa = 11.84SQEE332 pKa = 3.89GKK334 pKa = 9.83SRR336 pKa = 11.84RR337 pKa = 11.84APGQEE342 pKa = 3.32RR343 pKa = 11.84RR344 pKa = 11.84ASGVWRR350 pKa = 11.84RR351 pKa = 11.84HH352 pKa = 5.2ADD354 pKa = 3.16SGGGPGGAGAHH365 pKa = 6.71HH366 pKa = 6.53GRR368 pKa = 11.84DD369 pKa = 3.52PRR371 pKa = 11.84ATSMAGGRR379 pKa = 11.84RR380 pKa = 11.84AGLRR384 pKa = 11.84QCRR387 pKa = 11.84CAAWAGAVGRR397 pKa = 11.84GAGGSRR403 pKa = 11.84CSGG406 pKa = 3.18
Molecular weight: 44.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5851458 |
50 |
7188 |
505.8 |
55.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.791 ± 0.019 | 1.249 ± 0.008 |
5.743 ± 0.014 | 6.22 ± 0.02 |
3.677 ± 0.012 | 6.917 ± 0.021 |
2.426 ± 0.009 | 4.855 ± 0.014 |
4.411 ± 0.021 | 9.158 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.06 ± 0.009 | 3.584 ± 0.011 |
6.112 ± 0.022 | 3.918 ± 0.016 |
6.225 ± 0.021 | 8.171 ± 0.024 |
5.979 ± 0.016 | 6.199 ± 0.015 |
1.487 ± 0.008 | 2.818 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
