
gamma proteobacterium HTCC5015
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; unclassified Gammaproteobacteria
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
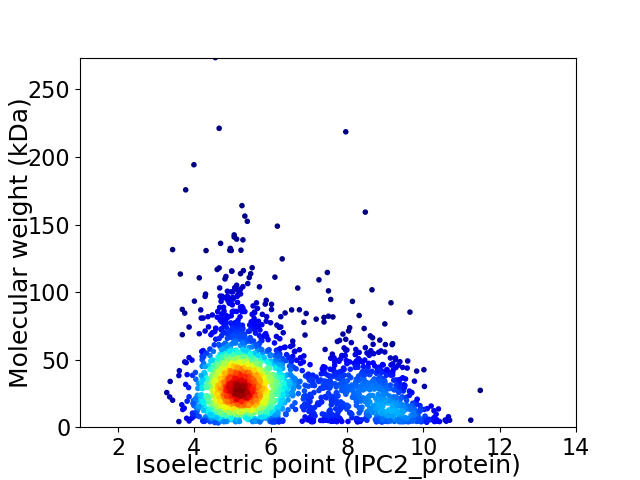
Virtual 2D-PAGE plot for 2449 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B5JUG5|B5JUG5_9GAMM Transposase OS=gamma proteobacterium HTCC5015 OX=391615 GN=GP5015_1446 PE=3 SV=1
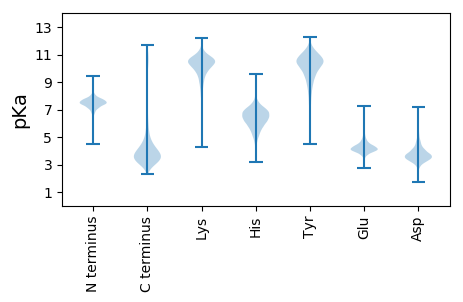
MM1 pKa = 7.38KK2 pKa = 10.49KK3 pKa = 10.01EE4 pKa = 3.88FARR7 pKa = 11.84SGKK10 pKa = 9.57HH11 pKa = 4.98VFMSLLLLLCYY22 pKa = 10.41SQAATANSTGISGYY36 pKa = 10.01SGEE39 pKa = 4.68SGSSCVSCHH48 pKa = 6.08NGNVYY53 pKa = 10.67NYY55 pKa = 9.91NSVISGDD62 pKa = 3.6STLTPNSSSSYY73 pKa = 8.3TFSLNRR79 pKa = 11.84VAGSTAGAAGFNLSADD95 pKa = 3.66NGTLSSGSGSKK106 pKa = 9.85ILNSEE111 pKa = 4.16LTHH114 pKa = 6.12SAKK117 pKa = 9.2KK118 pKa = 9.02TSNFNWSFNWTAPSADD134 pKa = 3.17GVYY137 pKa = 9.87TLYY140 pKa = 11.2ACGNPVDD147 pKa = 3.98NDD149 pKa = 3.82GVGLSDD155 pKa = 4.21TGDD158 pKa = 3.58GPADD162 pKa = 3.43CTEE165 pKa = 3.96FDD167 pKa = 3.6ITVNTPPTANSDD179 pKa = 3.68SSSLSEE185 pKa = 4.83DD186 pKa = 2.94GSYY189 pKa = 11.0DD190 pKa = 3.46YY191 pKa = 11.24VYY193 pKa = 10.5PLSNDD198 pKa = 2.86SSGDD202 pKa = 3.55VGDD205 pKa = 3.93TKK207 pKa = 11.38SLIGVCATSTLDD219 pKa = 3.49YY220 pKa = 10.09YY221 pKa = 11.6CNSTSYY227 pKa = 10.44TPSQGSFYY235 pKa = 11.09VAGSYY240 pKa = 9.2VRR242 pKa = 11.84YY243 pKa = 9.79LPAANYY249 pKa = 10.53NGTFSIKK256 pKa = 10.77YY257 pKa = 9.91KK258 pKa = 9.75MQDD261 pKa = 2.42SYY263 pKa = 12.16GDD265 pKa = 3.25VDD267 pKa = 3.96TANITYY273 pKa = 8.29TVSSVNDD280 pKa = 3.72DD281 pKa = 3.81PVVSGVDD288 pKa = 3.23TTLSYY293 pKa = 11.38TEE295 pKa = 4.11GQGARR300 pKa = 11.84TLEE303 pKa = 4.1SSINLSDD310 pKa = 4.19VDD312 pKa = 3.71HH313 pKa = 6.43TQLNRR318 pKa = 11.84AEE320 pKa = 4.27VEE322 pKa = 3.57ISGNYY327 pKa = 9.28VSGQDD332 pKa = 3.24VLAASNCSGLSCSFNSTNGTLTLFGTASIATYY364 pKa = 10.69QNALEE369 pKa = 4.32SVTYY373 pKa = 10.82ANTSEE378 pKa = 4.53DD379 pKa = 3.69PSTATRR385 pKa = 11.84TVRR388 pKa = 11.84IRR390 pKa = 11.84VRR392 pKa = 11.84DD393 pKa = 3.4TAYY396 pKa = 10.46AYY398 pKa = 11.13SNYY401 pKa = 9.12DD402 pKa = 3.1TMNITVSAEE411 pKa = 3.26NDD413 pKa = 3.4APVVSGFDD421 pKa = 3.07TTLNYY426 pKa = 9.95TEE428 pKa = 4.73NDD430 pKa = 3.53GARR433 pKa = 11.84SLEE436 pKa = 4.24SSVSLNDD443 pKa = 3.39IDD445 pKa = 5.35SSQLNRR451 pKa = 11.84AEE453 pKa = 3.73IQIYY457 pKa = 9.59SGYY460 pKa = 10.44INGEE464 pKa = 4.1DD465 pKa = 3.3VLAAGNCSALSCSFNTGTGTYY486 pKa = 9.46VFSGNASLATYY497 pKa = 8.92EE498 pKa = 4.25AVIEE502 pKa = 4.27SLTYY506 pKa = 9.86TNQSEE511 pKa = 4.55APSTSPRR518 pKa = 11.84AVRR521 pKa = 11.84LRR523 pKa = 11.84VRR525 pKa = 11.84DD526 pKa = 3.51TDD528 pKa = 3.45NDD530 pKa = 3.68YY531 pKa = 11.82SNGDD535 pKa = 3.74TLSIAVTAVEE545 pKa = 5.0DD546 pKa = 3.94PADD549 pKa = 3.96FDD551 pKa = 4.32SPVYY555 pKa = 10.44EE556 pKa = 4.33RR557 pKa = 11.84IGVGTSSSTTQNIAEE572 pKa = 4.53DD573 pKa = 3.85NQLQLQFAANDD584 pKa = 3.91PEE586 pKa = 4.62GVSTEE591 pKa = 4.04FNLVSTNPVLTGPEE605 pKa = 3.94PLISVSSDD613 pKa = 2.65GLLSWTPDD621 pKa = 3.06GTRR624 pKa = 11.84SSVQAVIGVNDD635 pKa = 4.63NIGSSPDD642 pKa = 3.22NTFSITLTVTGSNDD656 pKa = 3.03APVITEE662 pKa = 4.01GASINVNMDD671 pKa = 2.9EE672 pKa = 4.78DD673 pKa = 4.16ASPTAFNLTLNASDD687 pKa = 5.33LEE689 pKa = 4.29NDD691 pKa = 3.44TLTWSIVSQASKK703 pKa = 9.52GTASTSGTGNSKK715 pKa = 10.91AILYY719 pKa = 8.82TPDD722 pKa = 3.46LNEE725 pKa = 4.32NGSDD729 pKa = 3.26AFVVRR734 pKa = 11.84VTDD737 pKa = 4.42GSLSDD742 pKa = 5.46DD743 pKa = 3.0ITVNVNIAPQADD755 pKa = 3.67APVITDD761 pKa = 3.59GKK763 pKa = 10.12AITVAMSEE771 pKa = 4.69DD772 pKa = 3.76GSPTPFEE779 pKa = 4.35LVLNATDD786 pKa = 4.28PDD788 pKa = 4.44AGDD791 pKa = 3.52TLTWRR796 pKa = 11.84ISQAASRR803 pKa = 11.84GATSIPGATRR813 pKa = 11.84AGQPIVYY820 pKa = 9.44KK821 pKa = 10.91PNDD824 pKa = 3.81DD825 pKa = 3.97YY826 pKa = 11.65FGSDD830 pKa = 2.85QFTVSVSDD838 pKa = 3.64GRR840 pKa = 11.84LSDD843 pKa = 4.49SIDD846 pKa = 3.27VTVNIAPVNDD856 pKa = 3.47NPVIQTIGTLPAATEE871 pKa = 3.85DD872 pKa = 3.28QLYY875 pKa = 9.49QFVVAVSDD883 pKa = 3.68VDD885 pKa = 3.65HH886 pKa = 6.82SVPEE890 pKa = 4.14DD891 pKa = 3.68LRR893 pKa = 11.84FSLNNAPDD901 pKa = 3.56GMQINATGQVSWTPSEE917 pKa = 4.23GVSGSGKK924 pKa = 9.27VGVRR928 pKa = 11.84VEE930 pKa = 4.5DD931 pKa = 3.48SAGGIDD937 pKa = 3.2AAMFSVSVSAVNDD950 pKa = 4.22GPSINSTPPNTATEE964 pKa = 4.42GEE966 pKa = 4.4LYY968 pKa = 10.22EE969 pKa = 4.25YY970 pKa = 9.52TLQVSDD976 pKa = 5.14PDD978 pKa = 4.07DD979 pKa = 5.52ANDD982 pKa = 4.29GSGALNFSLSNPPAGMNISNTGEE1005 pKa = 4.13ITWTPVQGDD1014 pKa = 4.05ANGSSQVYY1022 pKa = 10.22NFTASVKK1029 pKa = 10.87DD1030 pKa = 3.56GGEE1033 pKa = 4.33DD1034 pKa = 3.62GATTAVQAIQITVAIPDD1051 pKa = 3.69ADD1053 pKa = 3.86GDD1055 pKa = 4.28TVADD1059 pKa = 4.11YY1060 pKa = 11.22EE1061 pKa = 4.8DD1062 pKa = 4.01NCPAVANTDD1071 pKa = 3.53QANNDD1076 pKa = 3.21ADD1078 pKa = 4.67AEE1080 pKa = 4.4GDD1082 pKa = 3.89LCDD1085 pKa = 6.31DD1086 pKa = 5.43DD1087 pKa = 6.7DD1088 pKa = 6.98DD1089 pKa = 6.01NDD1091 pKa = 3.89GMPDD1095 pKa = 3.72DD1096 pKa = 5.28FEE1098 pKa = 5.53VDD1100 pKa = 3.31NDD1102 pKa = 4.08FDD1104 pKa = 4.95PLDD1107 pKa = 4.17EE1108 pKa = 5.51SDD1110 pKa = 3.84AASDD1114 pKa = 3.7RR1115 pKa = 11.84DD1116 pKa = 3.44GDD1118 pKa = 4.38GISNLDD1124 pKa = 3.53EE1125 pKa = 4.31YY1126 pKa = 11.64LNNTDD1131 pKa = 4.04PSTDD1135 pKa = 2.98SVAPVITLAPEE1146 pKa = 4.0EE1147 pKa = 4.18VSRR1150 pKa = 11.84NAKK1153 pKa = 10.08GLLTPVEE1160 pKa = 4.57HH1161 pKa = 6.53SASAEE1166 pKa = 3.93DD1167 pKa = 5.38AIDD1170 pKa = 4.24GPVAVTADD1178 pKa = 3.22KK1179 pKa = 10.47TGPFASGHH1187 pKa = 6.14HH1188 pKa = 5.26VVTWQASDD1196 pKa = 3.22TSKK1199 pKa = 11.34NLAQSDD1205 pKa = 4.33QIIDD1209 pKa = 3.81IYY1211 pKa = 10.36PRR1213 pKa = 11.84VYY1215 pKa = 10.46VSPSQTVGEE1224 pKa = 4.31GQVVNVPIHH1233 pKa = 5.03ITGNAVSYY1241 pKa = 9.86PVTVYY1246 pKa = 10.58YY1247 pKa = 10.77SIDD1250 pKa = 3.55GATDD1254 pKa = 3.3SSDD1257 pKa = 3.32HH1258 pKa = 6.95DD1259 pKa = 4.35LVAGQIDD1266 pKa = 4.07ITSGHH1271 pKa = 5.2VSEE1274 pKa = 5.16LTFNTLADD1282 pKa = 3.84TLNEE1286 pKa = 4.02GSEE1289 pKa = 4.29SLNIRR1294 pKa = 11.84LTAADD1299 pKa = 4.13GASLSSAVNHH1309 pKa = 6.37QITITDD1315 pKa = 4.2LPAPPRR1321 pKa = 11.84INLLAAQDD1329 pKa = 3.71GHH1331 pKa = 5.06STRR1334 pKa = 11.84TIYY1337 pKa = 10.83LDD1339 pKa = 2.94QGLVHH1344 pKa = 6.63FSANASDD1351 pKa = 4.33PNGDD1355 pKa = 4.0TLSHH1359 pKa = 6.46TWTASISGINTISGGGSNDD1378 pKa = 3.52DD1379 pKa = 3.25HH1380 pKa = 8.15WEE1382 pKa = 3.82IDD1384 pKa = 3.65PQNLVAGQSINIRR1397 pKa = 11.84LSTHH1401 pKa = 6.85DD1402 pKa = 3.58GRR1404 pKa = 11.84HH1405 pKa = 4.89TAVQSLSLGIKK1416 pKa = 10.29DD1417 pKa = 4.25SAPLLDD1423 pKa = 4.57AFEE1426 pKa = 5.91DD1427 pKa = 3.93SDD1429 pKa = 5.18GDD1431 pKa = 4.42GIDD1434 pKa = 4.53DD1435 pKa = 4.65AAEE1438 pKa = 4.15GWGDD1442 pKa = 3.62SDD1444 pKa = 4.61GDD1446 pKa = 4.16GIANYY1451 pKa = 10.41LDD1453 pKa = 3.73STPLSSALQTSTGDD1467 pKa = 3.69FEE1469 pKa = 5.32TSLQQAIQTDD1479 pKa = 3.61PGLTLKK1485 pKa = 10.83LGSHH1489 pKa = 7.13ASAQGASKK1497 pKa = 10.94ASIAANSLLQTGNEE1511 pKa = 4.24DD1512 pKa = 3.45YY1513 pKa = 11.26QLIGQAFDD1521 pKa = 4.21FEE1523 pKa = 4.53IHH1525 pKa = 6.73GLDD1528 pKa = 3.28TSLTQAHH1535 pKa = 5.5VVIPLAVRR1543 pKa = 11.84IPSDD1547 pKa = 3.41DD1548 pKa = 3.46AVVRR1552 pKa = 11.84KK1553 pKa = 7.31YY1554 pKa = 10.56HH1555 pKa = 6.88PEE1557 pKa = 3.97HH1558 pKa = 6.47GWRR1561 pKa = 11.84AFNSSEE1567 pKa = 3.57GDD1569 pKa = 3.67TIHH1572 pKa = 7.23SLRR1575 pKa = 11.84SPDD1578 pKa = 3.54GTCPEE1583 pKa = 4.97PGSEE1587 pKa = 4.05QYY1589 pKa = 10.47QEE1591 pKa = 4.0GLAIFANCLQLTLTDD1606 pKa = 4.57GGPNDD1611 pKa = 3.71TDD1613 pKa = 3.7GVRR1616 pKa = 11.84NGIIVDD1622 pKa = 3.99PSAIAVPKK1630 pKa = 10.33QSNSSNDD1637 pKa = 3.21SSNEE1641 pKa = 3.79SAPRR1645 pKa = 11.84AGGSSGLSPLFFLWLLLISFAAARR1669 pKa = 11.84RR1670 pKa = 11.84RR1671 pKa = 11.84KK1672 pKa = 10.02ASIQITPSHH1681 pKa = 6.34
MM1 pKa = 7.38KK2 pKa = 10.49KK3 pKa = 10.01EE4 pKa = 3.88FARR7 pKa = 11.84SGKK10 pKa = 9.57HH11 pKa = 4.98VFMSLLLLLCYY22 pKa = 10.41SQAATANSTGISGYY36 pKa = 10.01SGEE39 pKa = 4.68SGSSCVSCHH48 pKa = 6.08NGNVYY53 pKa = 10.67NYY55 pKa = 9.91NSVISGDD62 pKa = 3.6STLTPNSSSSYY73 pKa = 8.3TFSLNRR79 pKa = 11.84VAGSTAGAAGFNLSADD95 pKa = 3.66NGTLSSGSGSKK106 pKa = 9.85ILNSEE111 pKa = 4.16LTHH114 pKa = 6.12SAKK117 pKa = 9.2KK118 pKa = 9.02TSNFNWSFNWTAPSADD134 pKa = 3.17GVYY137 pKa = 9.87TLYY140 pKa = 11.2ACGNPVDD147 pKa = 3.98NDD149 pKa = 3.82GVGLSDD155 pKa = 4.21TGDD158 pKa = 3.58GPADD162 pKa = 3.43CTEE165 pKa = 3.96FDD167 pKa = 3.6ITVNTPPTANSDD179 pKa = 3.68SSSLSEE185 pKa = 4.83DD186 pKa = 2.94GSYY189 pKa = 11.0DD190 pKa = 3.46YY191 pKa = 11.24VYY193 pKa = 10.5PLSNDD198 pKa = 2.86SSGDD202 pKa = 3.55VGDD205 pKa = 3.93TKK207 pKa = 11.38SLIGVCATSTLDD219 pKa = 3.49YY220 pKa = 10.09YY221 pKa = 11.6CNSTSYY227 pKa = 10.44TPSQGSFYY235 pKa = 11.09VAGSYY240 pKa = 9.2VRR242 pKa = 11.84YY243 pKa = 9.79LPAANYY249 pKa = 10.53NGTFSIKK256 pKa = 10.77YY257 pKa = 9.91KK258 pKa = 9.75MQDD261 pKa = 2.42SYY263 pKa = 12.16GDD265 pKa = 3.25VDD267 pKa = 3.96TANITYY273 pKa = 8.29TVSSVNDD280 pKa = 3.72DD281 pKa = 3.81PVVSGVDD288 pKa = 3.23TTLSYY293 pKa = 11.38TEE295 pKa = 4.11GQGARR300 pKa = 11.84TLEE303 pKa = 4.1SSINLSDD310 pKa = 4.19VDD312 pKa = 3.71HH313 pKa = 6.43TQLNRR318 pKa = 11.84AEE320 pKa = 4.27VEE322 pKa = 3.57ISGNYY327 pKa = 9.28VSGQDD332 pKa = 3.24VLAASNCSGLSCSFNSTNGTLTLFGTASIATYY364 pKa = 10.69QNALEE369 pKa = 4.32SVTYY373 pKa = 10.82ANTSEE378 pKa = 4.53DD379 pKa = 3.69PSTATRR385 pKa = 11.84TVRR388 pKa = 11.84IRR390 pKa = 11.84VRR392 pKa = 11.84DD393 pKa = 3.4TAYY396 pKa = 10.46AYY398 pKa = 11.13SNYY401 pKa = 9.12DD402 pKa = 3.1TMNITVSAEE411 pKa = 3.26NDD413 pKa = 3.4APVVSGFDD421 pKa = 3.07TTLNYY426 pKa = 9.95TEE428 pKa = 4.73NDD430 pKa = 3.53GARR433 pKa = 11.84SLEE436 pKa = 4.24SSVSLNDD443 pKa = 3.39IDD445 pKa = 5.35SSQLNRR451 pKa = 11.84AEE453 pKa = 3.73IQIYY457 pKa = 9.59SGYY460 pKa = 10.44INGEE464 pKa = 4.1DD465 pKa = 3.3VLAAGNCSALSCSFNTGTGTYY486 pKa = 9.46VFSGNASLATYY497 pKa = 8.92EE498 pKa = 4.25AVIEE502 pKa = 4.27SLTYY506 pKa = 9.86TNQSEE511 pKa = 4.55APSTSPRR518 pKa = 11.84AVRR521 pKa = 11.84LRR523 pKa = 11.84VRR525 pKa = 11.84DD526 pKa = 3.51TDD528 pKa = 3.45NDD530 pKa = 3.68YY531 pKa = 11.82SNGDD535 pKa = 3.74TLSIAVTAVEE545 pKa = 5.0DD546 pKa = 3.94PADD549 pKa = 3.96FDD551 pKa = 4.32SPVYY555 pKa = 10.44EE556 pKa = 4.33RR557 pKa = 11.84IGVGTSSSTTQNIAEE572 pKa = 4.53DD573 pKa = 3.85NQLQLQFAANDD584 pKa = 3.91PEE586 pKa = 4.62GVSTEE591 pKa = 4.04FNLVSTNPVLTGPEE605 pKa = 3.94PLISVSSDD613 pKa = 2.65GLLSWTPDD621 pKa = 3.06GTRR624 pKa = 11.84SSVQAVIGVNDD635 pKa = 4.63NIGSSPDD642 pKa = 3.22NTFSITLTVTGSNDD656 pKa = 3.03APVITEE662 pKa = 4.01GASINVNMDD671 pKa = 2.9EE672 pKa = 4.78DD673 pKa = 4.16ASPTAFNLTLNASDD687 pKa = 5.33LEE689 pKa = 4.29NDD691 pKa = 3.44TLTWSIVSQASKK703 pKa = 9.52GTASTSGTGNSKK715 pKa = 10.91AILYY719 pKa = 8.82TPDD722 pKa = 3.46LNEE725 pKa = 4.32NGSDD729 pKa = 3.26AFVVRR734 pKa = 11.84VTDD737 pKa = 4.42GSLSDD742 pKa = 5.46DD743 pKa = 3.0ITVNVNIAPQADD755 pKa = 3.67APVITDD761 pKa = 3.59GKK763 pKa = 10.12AITVAMSEE771 pKa = 4.69DD772 pKa = 3.76GSPTPFEE779 pKa = 4.35LVLNATDD786 pKa = 4.28PDD788 pKa = 4.44AGDD791 pKa = 3.52TLTWRR796 pKa = 11.84ISQAASRR803 pKa = 11.84GATSIPGATRR813 pKa = 11.84AGQPIVYY820 pKa = 9.44KK821 pKa = 10.91PNDD824 pKa = 3.81DD825 pKa = 3.97YY826 pKa = 11.65FGSDD830 pKa = 2.85QFTVSVSDD838 pKa = 3.64GRR840 pKa = 11.84LSDD843 pKa = 4.49SIDD846 pKa = 3.27VTVNIAPVNDD856 pKa = 3.47NPVIQTIGTLPAATEE871 pKa = 3.85DD872 pKa = 3.28QLYY875 pKa = 9.49QFVVAVSDD883 pKa = 3.68VDD885 pKa = 3.65HH886 pKa = 6.82SVPEE890 pKa = 4.14DD891 pKa = 3.68LRR893 pKa = 11.84FSLNNAPDD901 pKa = 3.56GMQINATGQVSWTPSEE917 pKa = 4.23GVSGSGKK924 pKa = 9.27VGVRR928 pKa = 11.84VEE930 pKa = 4.5DD931 pKa = 3.48SAGGIDD937 pKa = 3.2AAMFSVSVSAVNDD950 pKa = 4.22GPSINSTPPNTATEE964 pKa = 4.42GEE966 pKa = 4.4LYY968 pKa = 10.22EE969 pKa = 4.25YY970 pKa = 9.52TLQVSDD976 pKa = 5.14PDD978 pKa = 4.07DD979 pKa = 5.52ANDD982 pKa = 4.29GSGALNFSLSNPPAGMNISNTGEE1005 pKa = 4.13ITWTPVQGDD1014 pKa = 4.05ANGSSQVYY1022 pKa = 10.22NFTASVKK1029 pKa = 10.87DD1030 pKa = 3.56GGEE1033 pKa = 4.33DD1034 pKa = 3.62GATTAVQAIQITVAIPDD1051 pKa = 3.69ADD1053 pKa = 3.86GDD1055 pKa = 4.28TVADD1059 pKa = 4.11YY1060 pKa = 11.22EE1061 pKa = 4.8DD1062 pKa = 4.01NCPAVANTDD1071 pKa = 3.53QANNDD1076 pKa = 3.21ADD1078 pKa = 4.67AEE1080 pKa = 4.4GDD1082 pKa = 3.89LCDD1085 pKa = 6.31DD1086 pKa = 5.43DD1087 pKa = 6.7DD1088 pKa = 6.98DD1089 pKa = 6.01NDD1091 pKa = 3.89GMPDD1095 pKa = 3.72DD1096 pKa = 5.28FEE1098 pKa = 5.53VDD1100 pKa = 3.31NDD1102 pKa = 4.08FDD1104 pKa = 4.95PLDD1107 pKa = 4.17EE1108 pKa = 5.51SDD1110 pKa = 3.84AASDD1114 pKa = 3.7RR1115 pKa = 11.84DD1116 pKa = 3.44GDD1118 pKa = 4.38GISNLDD1124 pKa = 3.53EE1125 pKa = 4.31YY1126 pKa = 11.64LNNTDD1131 pKa = 4.04PSTDD1135 pKa = 2.98SVAPVITLAPEE1146 pKa = 4.0EE1147 pKa = 4.18VSRR1150 pKa = 11.84NAKK1153 pKa = 10.08GLLTPVEE1160 pKa = 4.57HH1161 pKa = 6.53SASAEE1166 pKa = 3.93DD1167 pKa = 5.38AIDD1170 pKa = 4.24GPVAVTADD1178 pKa = 3.22KK1179 pKa = 10.47TGPFASGHH1187 pKa = 6.14HH1188 pKa = 5.26VVTWQASDD1196 pKa = 3.22TSKK1199 pKa = 11.34NLAQSDD1205 pKa = 4.33QIIDD1209 pKa = 3.81IYY1211 pKa = 10.36PRR1213 pKa = 11.84VYY1215 pKa = 10.46VSPSQTVGEE1224 pKa = 4.31GQVVNVPIHH1233 pKa = 5.03ITGNAVSYY1241 pKa = 9.86PVTVYY1246 pKa = 10.58YY1247 pKa = 10.77SIDD1250 pKa = 3.55GATDD1254 pKa = 3.3SSDD1257 pKa = 3.32HH1258 pKa = 6.95DD1259 pKa = 4.35LVAGQIDD1266 pKa = 4.07ITSGHH1271 pKa = 5.2VSEE1274 pKa = 5.16LTFNTLADD1282 pKa = 3.84TLNEE1286 pKa = 4.02GSEE1289 pKa = 4.29SLNIRR1294 pKa = 11.84LTAADD1299 pKa = 4.13GASLSSAVNHH1309 pKa = 6.37QITITDD1315 pKa = 4.2LPAPPRR1321 pKa = 11.84INLLAAQDD1329 pKa = 3.71GHH1331 pKa = 5.06STRR1334 pKa = 11.84TIYY1337 pKa = 10.83LDD1339 pKa = 2.94QGLVHH1344 pKa = 6.63FSANASDD1351 pKa = 4.33PNGDD1355 pKa = 4.0TLSHH1359 pKa = 6.46TWTASISGINTISGGGSNDD1378 pKa = 3.52DD1379 pKa = 3.25HH1380 pKa = 8.15WEE1382 pKa = 3.82IDD1384 pKa = 3.65PQNLVAGQSINIRR1397 pKa = 11.84LSTHH1401 pKa = 6.85DD1402 pKa = 3.58GRR1404 pKa = 11.84HH1405 pKa = 4.89TAVQSLSLGIKK1416 pKa = 10.29DD1417 pKa = 4.25SAPLLDD1423 pKa = 4.57AFEE1426 pKa = 5.91DD1427 pKa = 3.93SDD1429 pKa = 5.18GDD1431 pKa = 4.42GIDD1434 pKa = 4.53DD1435 pKa = 4.65AAEE1438 pKa = 4.15GWGDD1442 pKa = 3.62SDD1444 pKa = 4.61GDD1446 pKa = 4.16GIANYY1451 pKa = 10.41LDD1453 pKa = 3.73STPLSSALQTSTGDD1467 pKa = 3.69FEE1469 pKa = 5.32TSLQQAIQTDD1479 pKa = 3.61PGLTLKK1485 pKa = 10.83LGSHH1489 pKa = 7.13ASAQGASKK1497 pKa = 10.94ASIAANSLLQTGNEE1511 pKa = 4.24DD1512 pKa = 3.45YY1513 pKa = 11.26QLIGQAFDD1521 pKa = 4.21FEE1523 pKa = 4.53IHH1525 pKa = 6.73GLDD1528 pKa = 3.28TSLTQAHH1535 pKa = 5.5VVIPLAVRR1543 pKa = 11.84IPSDD1547 pKa = 3.41DD1548 pKa = 3.46AVVRR1552 pKa = 11.84KK1553 pKa = 7.31YY1554 pKa = 10.56HH1555 pKa = 6.88PEE1557 pKa = 3.97HH1558 pKa = 6.47GWRR1561 pKa = 11.84AFNSSEE1567 pKa = 3.57GDD1569 pKa = 3.67TIHH1572 pKa = 7.23SLRR1575 pKa = 11.84SPDD1578 pKa = 3.54GTCPEE1583 pKa = 4.97PGSEE1587 pKa = 4.05QYY1589 pKa = 10.47QEE1591 pKa = 4.0GLAIFANCLQLTLTDD1606 pKa = 4.57GGPNDD1611 pKa = 3.71TDD1613 pKa = 3.7GVRR1616 pKa = 11.84NGIIVDD1622 pKa = 3.99PSAIAVPKK1630 pKa = 10.33QSNSSNDD1637 pKa = 3.21SSNEE1641 pKa = 3.79SAPRR1645 pKa = 11.84AGGSSGLSPLFFLWLLLISFAAARR1669 pKa = 11.84RR1670 pKa = 11.84RR1671 pKa = 11.84KK1672 pKa = 10.02ASIQITPSHH1681 pKa = 6.34
Molecular weight: 175.68 kDa
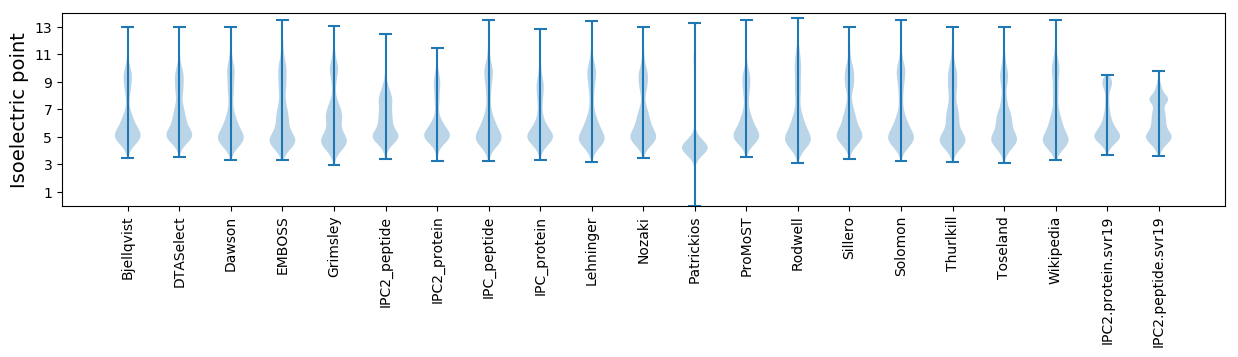
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B5JT67|B5JT67_9GAMM General secretion pathway protein F OS=gamma proteobacterium HTCC5015 OX=391615 GN=GP5015_2504 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.2RR14 pKa = 11.84THH16 pKa = 6.01GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.28GRR39 pKa = 11.84HH40 pKa = 5.14SLSAA44 pKa = 3.67
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.2RR14 pKa = 11.84THH16 pKa = 6.01GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.28GRR39 pKa = 11.84HH40 pKa = 5.14SLSAA44 pKa = 3.67
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
755730 |
30 |
2515 |
308.6 |
34.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.811 ± 0.065 | 0.971 ± 0.018 |
5.958 ± 0.041 | 6.696 ± 0.054 |
3.687 ± 0.032 | 7.434 ± 0.051 |
2.426 ± 0.03 | 5.247 ± 0.04 |
4.215 ± 0.048 | 10.33 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.49 ± 0.024 | 3.367 ± 0.034 |
4.3 ± 0.028 | 4.618 ± 0.04 |
6.053 ± 0.05 | 6.297 ± 0.043 |
4.866 ± 0.041 | 7.044 ± 0.045 |
1.354 ± 0.023 | 2.832 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
