
Capybara microvirus Cap1_SP_230
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
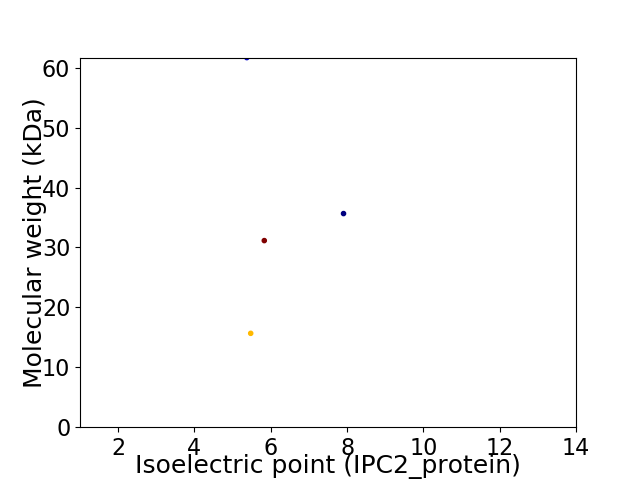
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5H5|A0A4P8W5H5_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_230 OX=2585416 PE=4 SV=1
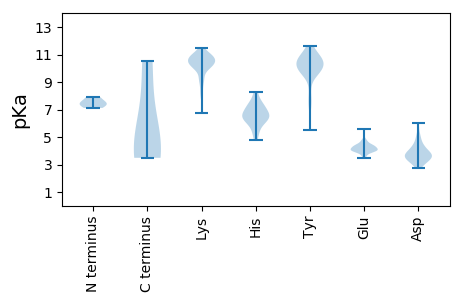
MM1 pKa = 7.9DD2 pKa = 4.59FSTRR6 pKa = 11.84YY7 pKa = 9.64RR8 pKa = 11.84PFKK11 pKa = 10.78AKK13 pKa = 10.2PCDD16 pKa = 3.61FFGDD20 pKa = 3.46EE21 pKa = 4.77RR22 pKa = 11.84ILVDD26 pKa = 3.73SSHH29 pKa = 7.16RR30 pKa = 11.84DD31 pKa = 3.23QVDD34 pKa = 2.77INNILKK40 pKa = 10.25RR41 pKa = 11.84YY42 pKa = 7.82RR43 pKa = 11.84QTGVLPPSLAQAVFADD59 pKa = 3.81TSIFTDD65 pKa = 3.61YY66 pKa = 10.71KK67 pKa = 10.88AALATIRR74 pKa = 11.84EE75 pKa = 4.38AEE77 pKa = 4.75DD78 pKa = 3.14IFASMPAEE86 pKa = 3.61IRR88 pKa = 11.84EE89 pKa = 4.32SCGYY93 pKa = 9.99EE94 pKa = 3.91PSEE97 pKa = 4.18LLAFMSRR104 pKa = 11.84PDD106 pKa = 4.2SKK108 pKa = 11.15PLIQKK113 pKa = 9.25YY114 pKa = 9.96FGGGVLPNQVAEE126 pKa = 4.49ANPAGISSPSPTKK139 pKa = 10.67KK140 pKa = 9.94EE141 pKa = 3.79
MM1 pKa = 7.9DD2 pKa = 4.59FSTRR6 pKa = 11.84YY7 pKa = 9.64RR8 pKa = 11.84PFKK11 pKa = 10.78AKK13 pKa = 10.2PCDD16 pKa = 3.61FFGDD20 pKa = 3.46EE21 pKa = 4.77RR22 pKa = 11.84ILVDD26 pKa = 3.73SSHH29 pKa = 7.16RR30 pKa = 11.84DD31 pKa = 3.23QVDD34 pKa = 2.77INNILKK40 pKa = 10.25RR41 pKa = 11.84YY42 pKa = 7.82RR43 pKa = 11.84QTGVLPPSLAQAVFADD59 pKa = 3.81TSIFTDD65 pKa = 3.61YY66 pKa = 10.71KK67 pKa = 10.88AALATIRR74 pKa = 11.84EE75 pKa = 4.38AEE77 pKa = 4.75DD78 pKa = 3.14IFASMPAEE86 pKa = 3.61IRR88 pKa = 11.84EE89 pKa = 4.32SCGYY93 pKa = 9.99EE94 pKa = 3.91PSEE97 pKa = 4.18LLAFMSRR104 pKa = 11.84PDD106 pKa = 4.2SKK108 pKa = 11.15PLIQKK113 pKa = 9.25YY114 pKa = 9.96FGGGVLPNQVAEE126 pKa = 4.49ANPAGISSPSPTKK139 pKa = 10.67KK140 pKa = 9.94EE141 pKa = 3.79
Molecular weight: 15.67 kDa
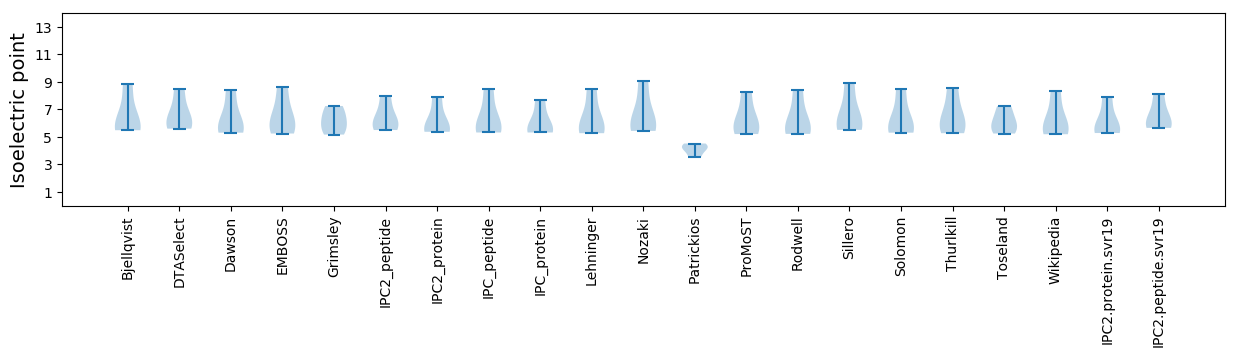
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W878|A0A4P8W878_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_230 OX=2585416 PE=4 SV=1
MM1 pKa = 7.44ICEE4 pKa = 4.31SPLQAFCLDD13 pKa = 3.83PSAQKK18 pKa = 10.62VCGIVDD24 pKa = 4.05FSKK27 pKa = 10.95LVDD30 pKa = 3.62GLTPRR35 pKa = 11.84IIFNSWLVNGAFPKK49 pKa = 10.45LCSLMEE55 pKa = 3.9QRR57 pKa = 11.84LCRR60 pKa = 11.84PVMLPCGKK68 pKa = 10.05CLACRR73 pKa = 11.84NRR75 pKa = 11.84HH76 pKa = 6.16LIDD79 pKa = 3.12VSLRR83 pKa = 11.84MQHH86 pKa = 6.08EE87 pKa = 5.55LITTVQPSTFLTLTYY102 pKa = 10.95NEE104 pKa = 4.08AHH106 pKa = 6.42SCNTLDD112 pKa = 4.11YY113 pKa = 11.56GDD115 pKa = 3.72IQSFLKK121 pKa = 10.54RR122 pKa = 11.84LRR124 pKa = 11.84TKK126 pKa = 10.44ALSPFRR132 pKa = 11.84FSCCGEE138 pKa = 3.71YY139 pKa = 10.95GKK141 pKa = 10.4LHH143 pKa = 6.24NRR145 pKa = 11.84PHH147 pKa = 5.49WHH149 pKa = 6.45FILYY153 pKa = 9.95GFYY156 pKa = 10.21PSDD159 pKa = 5.56GILQSQTRR167 pKa = 11.84GGNYY171 pKa = 9.29YY172 pKa = 10.75VSDD175 pKa = 4.8TISKK179 pKa = 8.12IWPFGFHH186 pKa = 6.73LFAPASAGACSYY198 pKa = 9.67LAKK201 pKa = 10.78YY202 pKa = 9.67LVKK205 pKa = 10.62QEE207 pKa = 4.66DD208 pKa = 4.82GIFKK212 pKa = 10.3QSLRR216 pKa = 11.84PGVGFEE222 pKa = 4.26YY223 pKa = 9.93YY224 pKa = 9.93RR225 pKa = 11.84KK226 pKa = 9.22YY227 pKa = 10.47HH228 pKa = 5.54VEE230 pKa = 4.44FYY232 pKa = 11.29SRR234 pKa = 11.84GFCVMNGFKK243 pKa = 10.78LPIPDD248 pKa = 3.54YY249 pKa = 10.53YY250 pKa = 11.43DD251 pKa = 5.21RR252 pKa = 11.84ILEE255 pKa = 4.29RR256 pKa = 11.84EE257 pKa = 4.18DD258 pKa = 2.96KK259 pKa = 11.0KK260 pKa = 10.93MYY262 pKa = 10.83EE263 pKa = 4.23EE264 pKa = 4.37VCKK267 pKa = 10.65KK268 pKa = 10.3RR269 pKa = 11.84LPCYY273 pKa = 10.35NIYY276 pKa = 10.62DD277 pKa = 3.76LSEE280 pKa = 3.97FLDD283 pKa = 4.02FHH285 pKa = 7.56RR286 pKa = 11.84HH287 pKa = 5.14DD288 pKa = 4.73AYY290 pKa = 10.72HH291 pKa = 6.67GKK293 pKa = 9.8ARR295 pKa = 11.84CLKK298 pKa = 10.42ISTQPEE304 pKa = 4.11KK305 pKa = 11.11GVLL308 pKa = 3.49
MM1 pKa = 7.44ICEE4 pKa = 4.31SPLQAFCLDD13 pKa = 3.83PSAQKK18 pKa = 10.62VCGIVDD24 pKa = 4.05FSKK27 pKa = 10.95LVDD30 pKa = 3.62GLTPRR35 pKa = 11.84IIFNSWLVNGAFPKK49 pKa = 10.45LCSLMEE55 pKa = 3.9QRR57 pKa = 11.84LCRR60 pKa = 11.84PVMLPCGKK68 pKa = 10.05CLACRR73 pKa = 11.84NRR75 pKa = 11.84HH76 pKa = 6.16LIDD79 pKa = 3.12VSLRR83 pKa = 11.84MQHH86 pKa = 6.08EE87 pKa = 5.55LITTVQPSTFLTLTYY102 pKa = 10.95NEE104 pKa = 4.08AHH106 pKa = 6.42SCNTLDD112 pKa = 4.11YY113 pKa = 11.56GDD115 pKa = 3.72IQSFLKK121 pKa = 10.54RR122 pKa = 11.84LRR124 pKa = 11.84TKK126 pKa = 10.44ALSPFRR132 pKa = 11.84FSCCGEE138 pKa = 3.71YY139 pKa = 10.95GKK141 pKa = 10.4LHH143 pKa = 6.24NRR145 pKa = 11.84PHH147 pKa = 5.49WHH149 pKa = 6.45FILYY153 pKa = 9.95GFYY156 pKa = 10.21PSDD159 pKa = 5.56GILQSQTRR167 pKa = 11.84GGNYY171 pKa = 9.29YY172 pKa = 10.75VSDD175 pKa = 4.8TISKK179 pKa = 8.12IWPFGFHH186 pKa = 6.73LFAPASAGACSYY198 pKa = 9.67LAKK201 pKa = 10.78YY202 pKa = 9.67LVKK205 pKa = 10.62QEE207 pKa = 4.66DD208 pKa = 4.82GIFKK212 pKa = 10.3QSLRR216 pKa = 11.84PGVGFEE222 pKa = 4.26YY223 pKa = 9.93YY224 pKa = 9.93RR225 pKa = 11.84KK226 pKa = 9.22YY227 pKa = 10.47HH228 pKa = 5.54VEE230 pKa = 4.44FYY232 pKa = 11.29SRR234 pKa = 11.84GFCVMNGFKK243 pKa = 10.78LPIPDD248 pKa = 3.54YY249 pKa = 10.53YY250 pKa = 11.43DD251 pKa = 5.21RR252 pKa = 11.84ILEE255 pKa = 4.29RR256 pKa = 11.84EE257 pKa = 4.18DD258 pKa = 2.96KK259 pKa = 11.0KK260 pKa = 10.93MYY262 pKa = 10.83EE263 pKa = 4.23EE264 pKa = 4.37VCKK267 pKa = 10.65KK268 pKa = 10.3RR269 pKa = 11.84LPCYY273 pKa = 10.35NIYY276 pKa = 10.62DD277 pKa = 3.76LSEE280 pKa = 3.97FLDD283 pKa = 4.02FHH285 pKa = 7.56RR286 pKa = 11.84HH287 pKa = 5.14DD288 pKa = 4.73AYY290 pKa = 10.72HH291 pKa = 6.67GKK293 pKa = 9.8ARR295 pKa = 11.84CLKK298 pKa = 10.42ISTQPEE304 pKa = 4.11KK305 pKa = 11.11GVLL308 pKa = 3.49
Molecular weight: 35.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1283 |
141 |
553 |
320.8 |
36.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.638 ± 2.093 | 1.637 ± 1.06 |
5.456 ± 0.429 | 6.002 ± 1.048 |
6.08 ± 0.965 | 6.859 ± 0.475 |
2.338 ± 0.371 | 3.663 ± 0.718 |
5.612 ± 1.34 | 9.119 ± 0.594 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.338 ± 0.343 | 3.897 ± 0.44 |
5.066 ± 0.85 | 4.521 ± 0.622 |
5.3 ± 0.497 | 8.496 ± 0.903 |
4.91 ± 0.92 | 5.846 ± 0.975 |
1.091 ± 0.177 | 4.131 ± 0.594 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
