
Arabidopsis lyrata subsp. lyrata (Lyre-leaved rock-cress)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis; Arabidopsis lyrata
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
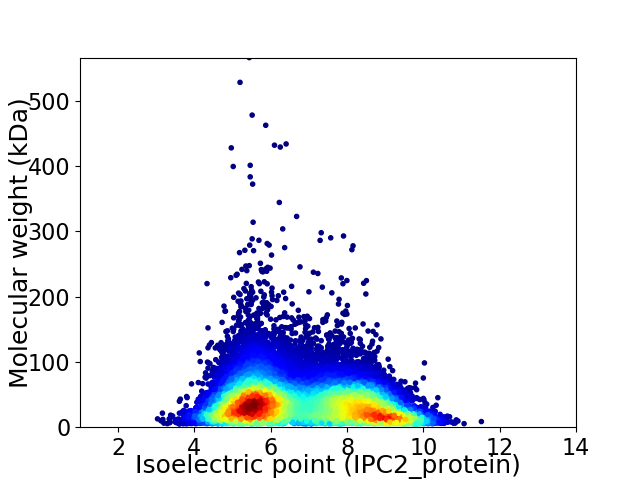
Virtual 2D-PAGE plot for 32113 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D7LQW4|D7LQW4_ARALL Expressed protein OS=Arabidopsis lyrata subsp. lyrata OX=81972 GN=ARALYDRAFT_904767 PE=4 SV=1
MM1 pKa = 7.43AVFVTSPDD9 pKa = 3.44EE10 pKa = 4.33NSDD13 pKa = 3.49VVYY16 pKa = 10.67SFSGYY21 pKa = 10.28SPASEE26 pKa = 5.16IVDD29 pKa = 3.79CYY31 pKa = 11.51LNGKK35 pKa = 8.97SPPKK39 pKa = 10.29LINSQSKK46 pKa = 10.62LGFWWEE52 pKa = 4.05DD53 pKa = 2.97PDD55 pKa = 4.75LYY57 pKa = 10.63RR58 pKa = 11.84YY59 pKa = 10.34CDD61 pKa = 4.52DD62 pKa = 4.42LSEE65 pKa = 4.97LNTIEE70 pKa = 3.97DD71 pKa = 2.99RR72 pKa = 11.84MMRR75 pKa = 11.84TRR77 pKa = 11.84KK78 pKa = 9.74HH79 pKa = 7.17LMDD82 pKa = 4.71CLEE85 pKa = 4.68KK86 pKa = 10.57KK87 pKa = 9.97EE88 pKa = 4.08KK89 pKa = 10.46SQFVSNSDD97 pKa = 3.42QNPNTDD103 pKa = 3.57EE104 pKa = 3.96IFNNGGSSSSSSQIASDD121 pKa = 4.19LGQNPSTSSPSSSQIVSFDD140 pKa = 3.52QNSYY144 pKa = 9.4TSLGKK149 pKa = 10.62LCGEE153 pKa = 4.6PSSQVTCYY161 pKa = 10.49DD162 pKa = 3.3QNPNFSVGEE171 pKa = 3.91YY172 pKa = 10.45SCDD175 pKa = 3.19QSRR178 pKa = 11.84YY179 pKa = 9.73LVNEE183 pKa = 3.89DD184 pKa = 4.19PGFVDD189 pKa = 4.8CLCEE193 pKa = 4.17TEE195 pKa = 4.36EE196 pKa = 4.43EE197 pKa = 4.37NNGMSLPQEE206 pKa = 4.25TQTPSLFTEE215 pKa = 4.7DD216 pKa = 3.17QSFWEE221 pKa = 4.22NLFKK225 pKa = 11.38DD226 pKa = 3.62EE227 pKa = 5.33DD228 pKa = 4.03NVFGLLNDD236 pKa = 3.69NLEE239 pKa = 4.36VPLQDD244 pKa = 3.37HH245 pKa = 6.61SSTNEE250 pKa = 3.72EE251 pKa = 3.78EE252 pKa = 4.32DD253 pKa = 3.38EE254 pKa = 4.26FMIDD258 pKa = 2.64ISEE261 pKa = 4.2FLSEE265 pKa = 4.25EE266 pKa = 4.52EE267 pKa = 3.99EE268 pKa = 4.53FEE270 pKa = 4.19WPLFF274 pKa = 3.95
MM1 pKa = 7.43AVFVTSPDD9 pKa = 3.44EE10 pKa = 4.33NSDD13 pKa = 3.49VVYY16 pKa = 10.67SFSGYY21 pKa = 10.28SPASEE26 pKa = 5.16IVDD29 pKa = 3.79CYY31 pKa = 11.51LNGKK35 pKa = 8.97SPPKK39 pKa = 10.29LINSQSKK46 pKa = 10.62LGFWWEE52 pKa = 4.05DD53 pKa = 2.97PDD55 pKa = 4.75LYY57 pKa = 10.63RR58 pKa = 11.84YY59 pKa = 10.34CDD61 pKa = 4.52DD62 pKa = 4.42LSEE65 pKa = 4.97LNTIEE70 pKa = 3.97DD71 pKa = 2.99RR72 pKa = 11.84MMRR75 pKa = 11.84TRR77 pKa = 11.84KK78 pKa = 9.74HH79 pKa = 7.17LMDD82 pKa = 4.71CLEE85 pKa = 4.68KK86 pKa = 10.57KK87 pKa = 9.97EE88 pKa = 4.08KK89 pKa = 10.46SQFVSNSDD97 pKa = 3.42QNPNTDD103 pKa = 3.57EE104 pKa = 3.96IFNNGGSSSSSSQIASDD121 pKa = 4.19LGQNPSTSSPSSSQIVSFDD140 pKa = 3.52QNSYY144 pKa = 9.4TSLGKK149 pKa = 10.62LCGEE153 pKa = 4.6PSSQVTCYY161 pKa = 10.49DD162 pKa = 3.3QNPNFSVGEE171 pKa = 3.91YY172 pKa = 10.45SCDD175 pKa = 3.19QSRR178 pKa = 11.84YY179 pKa = 9.73LVNEE183 pKa = 3.89DD184 pKa = 4.19PGFVDD189 pKa = 4.8CLCEE193 pKa = 4.17TEE195 pKa = 4.36EE196 pKa = 4.43EE197 pKa = 4.37NNGMSLPQEE206 pKa = 4.25TQTPSLFTEE215 pKa = 4.7DD216 pKa = 3.17QSFWEE221 pKa = 4.22NLFKK225 pKa = 11.38DD226 pKa = 3.62EE227 pKa = 5.33DD228 pKa = 4.03NVFGLLNDD236 pKa = 3.69NLEE239 pKa = 4.36VPLQDD244 pKa = 3.37HH245 pKa = 6.61SSTNEE250 pKa = 3.72EE251 pKa = 3.78EE252 pKa = 4.32DD253 pKa = 3.38EE254 pKa = 4.26FMIDD258 pKa = 2.64ISEE261 pKa = 4.2FLSEE265 pKa = 4.25EE266 pKa = 4.52EE267 pKa = 3.99EE268 pKa = 4.53FEE270 pKa = 4.19WPLFF274 pKa = 3.95
Molecular weight: 31.05 kDa
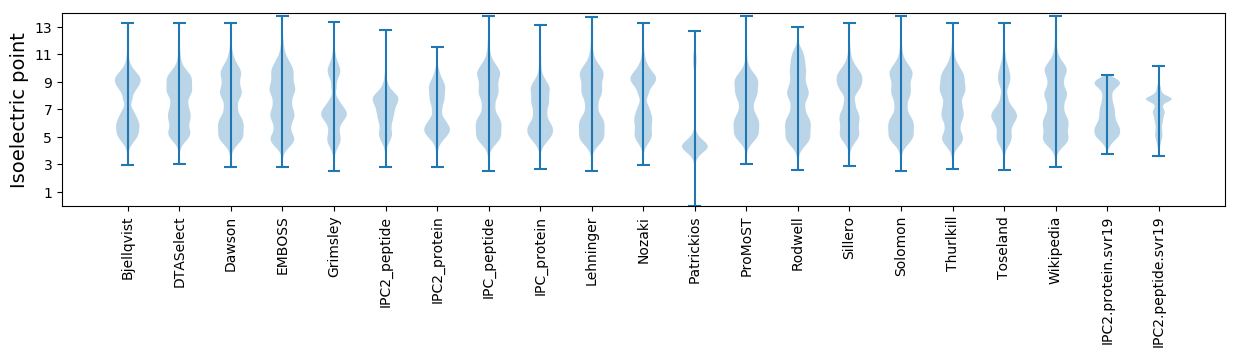
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D7MRS9|D7MRS9_ARALL Copper transporter OS=Arabidopsis lyrata subsp. lyrata OX=81972 GN=ARALYDRAFT_496052 PE=3 SV=1
LL1 pKa = 7.35HH2 pKa = 7.11LRR4 pKa = 11.84QLRR7 pKa = 11.84ILHH10 pKa = 5.79QNNIHH15 pKa = 5.95RR16 pKa = 11.84RR17 pKa = 11.84SRR19 pKa = 11.84SILRR23 pKa = 11.84QNNIHH28 pKa = 6.17RR29 pKa = 11.84RR30 pKa = 11.84SRR32 pKa = 11.84SILRR36 pKa = 11.84QNNIHH41 pKa = 6.04RR42 pKa = 11.84RR43 pKa = 11.84SRR45 pKa = 11.84STHH48 pKa = 4.61RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84TRR53 pKa = 11.84LTLIHH58 pKa = 6.67RR59 pKa = 11.84RR60 pKa = 11.84SKK62 pKa = 9.9LTLIRR67 pKa = 11.84RR68 pKa = 3.92
LL1 pKa = 7.35HH2 pKa = 7.11LRR4 pKa = 11.84QLRR7 pKa = 11.84ILHH10 pKa = 5.79QNNIHH15 pKa = 5.95RR16 pKa = 11.84RR17 pKa = 11.84SRR19 pKa = 11.84SILRR23 pKa = 11.84QNNIHH28 pKa = 6.17RR29 pKa = 11.84RR30 pKa = 11.84SRR32 pKa = 11.84SILRR36 pKa = 11.84QNNIHH41 pKa = 6.04RR42 pKa = 11.84RR43 pKa = 11.84SRR45 pKa = 11.84STHH48 pKa = 4.61RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84TRR53 pKa = 11.84LTLIHH58 pKa = 6.67RR59 pKa = 11.84RR60 pKa = 11.84SKK62 pKa = 9.9LTLIRR67 pKa = 11.84RR68 pKa = 3.92
Molecular weight: 8.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11683288 |
49 |
5090 |
363.8 |
40.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.294 ± 0.014 | 1.868 ± 0.008 |
5.37 ± 0.008 | 6.674 ± 0.016 |
4.349 ± 0.009 | 6.383 ± 0.015 |
2.261 ± 0.007 | 5.382 ± 0.011 |
6.357 ± 0.015 | 9.566 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.469 ± 0.005 | 4.389 ± 0.009 |
4.796 ± 0.013 | 3.445 ± 0.01 |
5.446 ± 0.01 | 9.064 ± 0.017 |
5.083 ± 0.008 | 6.691 ± 0.01 |
1.275 ± 0.005 | 2.838 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
