
Geobacter uraniireducens (strain Rf4) (Geobacter uraniumreducens)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfuromonadales; Geobacteraceae; Geobacter; Geobacter uraniireducens
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
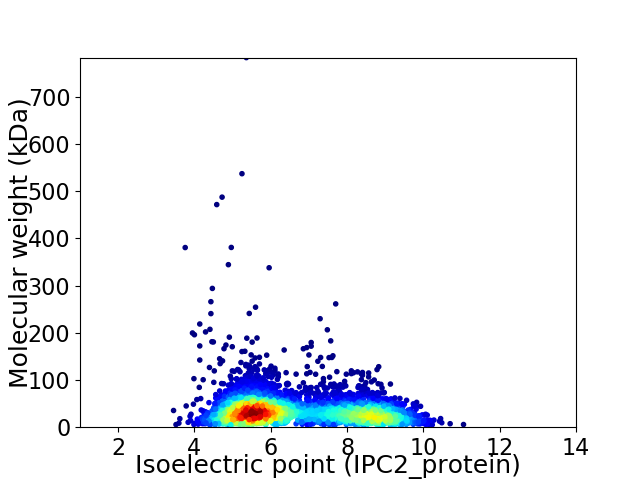
Virtual 2D-PAGE plot for 4255 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A5GBY5|A5GBY5_GEOUR TPR repeat-containing protein OS=Geobacter uraniireducens (strain Rf4) OX=351605 GN=Gura_0702 PE=4 SV=1
MM1 pKa = 7.69SIRR4 pKa = 11.84MTSAKK9 pKa = 9.64ILRR12 pKa = 11.84GRR14 pKa = 11.84ISFVATLLCMVLTQPLSSFAASVTLDD40 pKa = 3.56AADD43 pKa = 4.48DD44 pKa = 4.44VYY46 pKa = 11.07TDD48 pKa = 3.37SSRR51 pKa = 11.84IPTAGYY57 pKa = 6.23TTTDD61 pKa = 3.25MASVKK66 pKa = 10.06DD67 pKa = 4.05DD68 pKa = 3.74SMSKK72 pKa = 9.3IVNYY76 pKa = 10.17VGRR79 pKa = 11.84TPSISVLNGRR89 pKa = 11.84TVVKK93 pKa = 10.15FNLSGISGAINSASFKK109 pKa = 10.88FNVIDD114 pKa = 3.43IHH116 pKa = 5.77GTPILNLIKK125 pKa = 10.35VTDD128 pKa = 4.15NSWSEE133 pKa = 4.05QPVGASTFPTYY144 pKa = 10.81TDD146 pKa = 2.95TDD148 pKa = 4.63YY149 pKa = 10.34ITQNSTPYY157 pKa = 10.26SNQSVTTGWNNINVKK172 pKa = 10.5DD173 pKa = 4.0YY174 pKa = 10.8IQEE177 pKa = 4.39KK178 pKa = 9.93INAGSPYY185 pKa = 9.27VTIAMTGYY193 pKa = 9.09ATGATDD199 pKa = 4.6DD200 pKa = 5.29DD201 pKa = 5.02FDD203 pKa = 5.09FVSLNNGDD211 pKa = 4.2ALIPTLTVDD220 pKa = 3.44YY221 pKa = 8.69TPPPTVTGISPSSGPVAGGTSVTISGANFTSATAVTFGSTNATSYY266 pKa = 9.51TVNSATQITATSPAGSAGTVDD287 pKa = 3.17ITVTTAGGTSATGASDD303 pKa = 3.23QFTYY307 pKa = 10.41IAAPTVTSISPTSGPTDD324 pKa = 3.12GSTSVTITGTNFTGATTVTIGGAAATSVTVVNATTLTATTPSGTAGVRR372 pKa = 11.84DD373 pKa = 3.8VVVTTPGGTGTGTGLFTYY391 pKa = 9.97KK392 pKa = 10.5ASQTITFNSPGAQNFGTTPTLTAIASSGLTPTFTSSTTGVCTITSGGALTFATTGTCTINAGQAGDD458 pKa = 3.9STYY461 pKa = 11.23LAALPVSQSFSVAAVVPGAPTIGTATAGDD490 pKa = 3.9TQASVTFTAPASNGGASITGYY511 pKa = 8.18TATSNPGGVTGTCASSPCTVTGLTNGTAYY540 pKa = 9.71TFTVTATNSAGTGSASAASNSVTPAAAQTITFNNPGAQNFGIAPTLTATASSGLMVSFTSSTTGVCTVTAGGALTFVTAGTCTINADD627 pKa = 3.47QAGNGSFTAAPMVTRR642 pKa = 11.84SFAVNAVVPGAPTIGTATAGDD663 pKa = 3.94TQASITFSAPASNGGASITSYY684 pKa = 9.89TATSNPGGLTGTCASSPCTVTGLTNGTAYY713 pKa = 9.71TFTVTATNSAGTGSASAASNSVTPAAAQTITFNNPGAQNFGTSPTLTATATSSLTVTFTSSTTGVCTVTAGGALTFVTTGTCTINADD800 pKa = 3.47QAGNGSFLAATTVSRR815 pKa = 11.84SFTVIAVVPGAPTIGIATAGDD836 pKa = 3.76TQASVAFTAPVSNGGASITGYY857 pKa = 8.63TVTSNPGGLTSSGASSPITVTGLTNGTAYY886 pKa = 9.73TFTVTAHH893 pKa = 6.25NSAGTGSASSASNSVTPNPGPTVVNVAVPANGIYY927 pKa = 10.3KK928 pKa = 10.43AGSNLDD934 pKa = 3.79FTVTWDD940 pKa = 3.16SAATVTGTPRR950 pKa = 11.84IALLIGSAMVYY961 pKa = 8.82ATYY964 pKa = 10.66QSGSGTASTLFRR976 pKa = 11.84YY977 pKa = 7.35TVLPGQTDD985 pKa = 3.39TDD987 pKa = 5.02GITVGALSLNGGTIQNSSGTDD1008 pKa = 2.99ATLTLNSVASTVNVLVDD1025 pKa = 3.61TTAPTLSSIATSNPTHH1041 pKa = 6.87SGGTLTATANEE1052 pKa = 4.12NALGSWIAVSSSATAPTVAQVLAGADD1078 pKa = 3.57YY1079 pKa = 11.5GGVTVVAHH1087 pKa = 6.4GSGALSAGSAVSFSLSGLVAATSYY1111 pKa = 11.01DD1112 pKa = 3.33IYY1114 pKa = 11.2LAAQDD1119 pKa = 3.9AAGNPSAAVSSATLITTTTRR1139 pKa = 11.84VVTTSSDD1146 pKa = 3.4SGPGSLRR1153 pKa = 11.84QTIADD1158 pKa = 4.45ANPGDD1163 pKa = 4.82IILFDD1168 pKa = 4.7PSLSGQTVTIASPLVIAKK1186 pKa = 9.72DD1187 pKa = 3.76LSIGGYY1193 pKa = 7.5DD1194 pKa = 4.12ARR1196 pKa = 11.84PITINGGGTTRR1207 pKa = 11.84IFQVSGSTTFTLNYY1221 pKa = 8.15LTLTDD1226 pKa = 4.86GVATDD1231 pKa = 4.61GGAISDD1237 pKa = 4.05NVNATTFISLCTFSGNTATAAGGAISAAGTMSISDD1272 pKa = 3.59TLFAGNSAVQGGAIFNNNALSLVNVTLANNSANSGGGIYY1311 pKa = 10.33SSGGSTLVKK1320 pKa = 9.68NTTIAGNNATVQGGGIEE1337 pKa = 4.16IASGAVGFRR1346 pKa = 11.84NSIVAGNTSPSGPDD1360 pKa = 2.72ISGFATSLGYY1370 pKa = 10.94NLVKK1374 pKa = 9.99DD1375 pKa = 3.7TSGATFTVTTGDD1387 pKa = 3.54LTGQDD1392 pKa = 4.01PLLGPLADD1400 pKa = 3.62NGGPIKK1406 pKa = 10.12TMALLLGSPAIDD1418 pKa = 3.4SGACTGASATDD1429 pKa = 3.62QRR1431 pKa = 11.84SMPRR1435 pKa = 11.84PQNGLCDD1442 pKa = 3.27MGAYY1446 pKa = 9.27EE1447 pKa = 4.65RR1448 pKa = 11.84GVPAALTATGGTPQSAAIDD1467 pKa = 3.65AAFATPLKK1475 pKa = 10.96AKK1477 pKa = 9.25VTDD1480 pKa = 3.63SLGGVMEE1487 pKa = 5.42GISVTFAGPGSGADD1501 pKa = 3.13ITADD1505 pKa = 3.28GSVTTDD1511 pKa = 2.94AAGIASYY1518 pKa = 10.98GVTANGTAGAYY1529 pKa = 7.62TVTATVNALIANFNLTNDD1547 pKa = 4.18KK1548 pKa = 10.93ANQAITFNPPATATFGDD1565 pKa = 4.0APIALSATGGASGNPVIFSVASGPGSLNGATLTITGAGNIVVTASQAGNANYY1617 pKa = 9.97NAAPQVIRR1625 pKa = 11.84NIAIGKK1631 pKa = 8.53GAATIALGSLNATYY1645 pKa = 10.68DD1646 pKa = 3.61GTAKK1650 pKa = 10.63AVTATTTPAGLAVIVTYY1667 pKa = 10.45GGSSTPPTAAGSYY1680 pKa = 9.25PVAATIDD1687 pKa = 3.59DD1688 pKa = 3.95ANYY1691 pKa = 10.58SGTATGTLVIAYY1703 pKa = 8.37SATPPVLTISTLADD1717 pKa = 3.25GSVTNNATLNVSGQATSINGITSVTVNGAAVTLAADD1753 pKa = 4.04GTFSQAVTLQAGTNTVTTIATDD1775 pKa = 3.49NAGTTTTDD1783 pKa = 2.49SRR1785 pKa = 11.84TITLDD1790 pKa = 2.78TTAPVITITTPADD1803 pKa = 3.43NSTLAASSVTITGSVDD1819 pKa = 2.92KK1820 pKa = 10.79NATVQATANSGSPQSAAMTNNTFTVTLNLAGGSNTIVISATDD1862 pKa = 3.6LAGNSASVKK1871 pKa = 9.12RR1872 pKa = 11.84TIVSDD1877 pKa = 3.48TTKK1880 pKa = 9.41PTLAITNPSQDD1891 pKa = 2.88ITISEE1896 pKa = 4.25PALTISGTVTDD1907 pKa = 4.6ALTDD1911 pKa = 3.41VSVTITCDD1919 pKa = 2.83GKK1921 pKa = 9.18TYY1923 pKa = 10.05TPQVVDD1929 pKa = 4.49GAFQQQLTFVMAKK1942 pKa = 9.48QYY1944 pKa = 10.93AITVTATDD1952 pKa = 3.34QAGNSVTTQRR1962 pKa = 11.84NVIYY1966 pKa = 9.92ALSSLPSGDD1975 pKa = 3.15INGDD1979 pKa = 3.62GKK1981 pKa = 11.42VDD1983 pKa = 3.26IADD1986 pKa = 4.06ALLALQMAVGLITPTSAQLATGDD2009 pKa = 4.0VAPLSNGKK2017 pKa = 8.45PAPDD2021 pKa = 3.62GAIDD2025 pKa = 3.6IADD2028 pKa = 3.46AMLILEE2034 pKa = 4.79KK2035 pKa = 10.64AVGMLTWW2042 pKa = 4.01
MM1 pKa = 7.69SIRR4 pKa = 11.84MTSAKK9 pKa = 9.64ILRR12 pKa = 11.84GRR14 pKa = 11.84ISFVATLLCMVLTQPLSSFAASVTLDD40 pKa = 3.56AADD43 pKa = 4.48DD44 pKa = 4.44VYY46 pKa = 11.07TDD48 pKa = 3.37SSRR51 pKa = 11.84IPTAGYY57 pKa = 6.23TTTDD61 pKa = 3.25MASVKK66 pKa = 10.06DD67 pKa = 4.05DD68 pKa = 3.74SMSKK72 pKa = 9.3IVNYY76 pKa = 10.17VGRR79 pKa = 11.84TPSISVLNGRR89 pKa = 11.84TVVKK93 pKa = 10.15FNLSGISGAINSASFKK109 pKa = 10.88FNVIDD114 pKa = 3.43IHH116 pKa = 5.77GTPILNLIKK125 pKa = 10.35VTDD128 pKa = 4.15NSWSEE133 pKa = 4.05QPVGASTFPTYY144 pKa = 10.81TDD146 pKa = 2.95TDD148 pKa = 4.63YY149 pKa = 10.34ITQNSTPYY157 pKa = 10.26SNQSVTTGWNNINVKK172 pKa = 10.5DD173 pKa = 4.0YY174 pKa = 10.8IQEE177 pKa = 4.39KK178 pKa = 9.93INAGSPYY185 pKa = 9.27VTIAMTGYY193 pKa = 9.09ATGATDD199 pKa = 4.6DD200 pKa = 5.29DD201 pKa = 5.02FDD203 pKa = 5.09FVSLNNGDD211 pKa = 4.2ALIPTLTVDD220 pKa = 3.44YY221 pKa = 8.69TPPPTVTGISPSSGPVAGGTSVTISGANFTSATAVTFGSTNATSYY266 pKa = 9.51TVNSATQITATSPAGSAGTVDD287 pKa = 3.17ITVTTAGGTSATGASDD303 pKa = 3.23QFTYY307 pKa = 10.41IAAPTVTSISPTSGPTDD324 pKa = 3.12GSTSVTITGTNFTGATTVTIGGAAATSVTVVNATTLTATTPSGTAGVRR372 pKa = 11.84DD373 pKa = 3.8VVVTTPGGTGTGTGLFTYY391 pKa = 9.97KK392 pKa = 10.5ASQTITFNSPGAQNFGTTPTLTAIASSGLTPTFTSSTTGVCTITSGGALTFATTGTCTINAGQAGDD458 pKa = 3.9STYY461 pKa = 11.23LAALPVSQSFSVAAVVPGAPTIGTATAGDD490 pKa = 3.9TQASVTFTAPASNGGASITGYY511 pKa = 8.18TATSNPGGVTGTCASSPCTVTGLTNGTAYY540 pKa = 9.71TFTVTATNSAGTGSASAASNSVTPAAAQTITFNNPGAQNFGIAPTLTATASSGLMVSFTSSTTGVCTVTAGGALTFVTAGTCTINADD627 pKa = 3.47QAGNGSFTAAPMVTRR642 pKa = 11.84SFAVNAVVPGAPTIGTATAGDD663 pKa = 3.94TQASITFSAPASNGGASITSYY684 pKa = 9.89TATSNPGGLTGTCASSPCTVTGLTNGTAYY713 pKa = 9.71TFTVTATNSAGTGSASAASNSVTPAAAQTITFNNPGAQNFGTSPTLTATATSSLTVTFTSSTTGVCTVTAGGALTFVTTGTCTINADD800 pKa = 3.47QAGNGSFLAATTVSRR815 pKa = 11.84SFTVIAVVPGAPTIGIATAGDD836 pKa = 3.76TQASVAFTAPVSNGGASITGYY857 pKa = 8.63TVTSNPGGLTSSGASSPITVTGLTNGTAYY886 pKa = 9.73TFTVTAHH893 pKa = 6.25NSAGTGSASSASNSVTPNPGPTVVNVAVPANGIYY927 pKa = 10.3KK928 pKa = 10.43AGSNLDD934 pKa = 3.79FTVTWDD940 pKa = 3.16SAATVTGTPRR950 pKa = 11.84IALLIGSAMVYY961 pKa = 8.82ATYY964 pKa = 10.66QSGSGTASTLFRR976 pKa = 11.84YY977 pKa = 7.35TVLPGQTDD985 pKa = 3.39TDD987 pKa = 5.02GITVGALSLNGGTIQNSSGTDD1008 pKa = 2.99ATLTLNSVASTVNVLVDD1025 pKa = 3.61TTAPTLSSIATSNPTHH1041 pKa = 6.87SGGTLTATANEE1052 pKa = 4.12NALGSWIAVSSSATAPTVAQVLAGADD1078 pKa = 3.57YY1079 pKa = 11.5GGVTVVAHH1087 pKa = 6.4GSGALSAGSAVSFSLSGLVAATSYY1111 pKa = 11.01DD1112 pKa = 3.33IYY1114 pKa = 11.2LAAQDD1119 pKa = 3.9AAGNPSAAVSSATLITTTTRR1139 pKa = 11.84VVTTSSDD1146 pKa = 3.4SGPGSLRR1153 pKa = 11.84QTIADD1158 pKa = 4.45ANPGDD1163 pKa = 4.82IILFDD1168 pKa = 4.7PSLSGQTVTIASPLVIAKK1186 pKa = 9.72DD1187 pKa = 3.76LSIGGYY1193 pKa = 7.5DD1194 pKa = 4.12ARR1196 pKa = 11.84PITINGGGTTRR1207 pKa = 11.84IFQVSGSTTFTLNYY1221 pKa = 8.15LTLTDD1226 pKa = 4.86GVATDD1231 pKa = 4.61GGAISDD1237 pKa = 4.05NVNATTFISLCTFSGNTATAAGGAISAAGTMSISDD1272 pKa = 3.59TLFAGNSAVQGGAIFNNNALSLVNVTLANNSANSGGGIYY1311 pKa = 10.33SSGGSTLVKK1320 pKa = 9.68NTTIAGNNATVQGGGIEE1337 pKa = 4.16IASGAVGFRR1346 pKa = 11.84NSIVAGNTSPSGPDD1360 pKa = 2.72ISGFATSLGYY1370 pKa = 10.94NLVKK1374 pKa = 9.99DD1375 pKa = 3.7TSGATFTVTTGDD1387 pKa = 3.54LTGQDD1392 pKa = 4.01PLLGPLADD1400 pKa = 3.62NGGPIKK1406 pKa = 10.12TMALLLGSPAIDD1418 pKa = 3.4SGACTGASATDD1429 pKa = 3.62QRR1431 pKa = 11.84SMPRR1435 pKa = 11.84PQNGLCDD1442 pKa = 3.27MGAYY1446 pKa = 9.27EE1447 pKa = 4.65RR1448 pKa = 11.84GVPAALTATGGTPQSAAIDD1467 pKa = 3.65AAFATPLKK1475 pKa = 10.96AKK1477 pKa = 9.25VTDD1480 pKa = 3.63SLGGVMEE1487 pKa = 5.42GISVTFAGPGSGADD1501 pKa = 3.13ITADD1505 pKa = 3.28GSVTTDD1511 pKa = 2.94AAGIASYY1518 pKa = 10.98GVTANGTAGAYY1529 pKa = 7.62TVTATVNALIANFNLTNDD1547 pKa = 4.18KK1548 pKa = 10.93ANQAITFNPPATATFGDD1565 pKa = 4.0APIALSATGGASGNPVIFSVASGPGSLNGATLTITGAGNIVVTASQAGNANYY1617 pKa = 9.97NAAPQVIRR1625 pKa = 11.84NIAIGKK1631 pKa = 8.53GAATIALGSLNATYY1645 pKa = 10.68DD1646 pKa = 3.61GTAKK1650 pKa = 10.63AVTATTTPAGLAVIVTYY1667 pKa = 10.45GGSSTPPTAAGSYY1680 pKa = 9.25PVAATIDD1687 pKa = 3.59DD1688 pKa = 3.95ANYY1691 pKa = 10.58SGTATGTLVIAYY1703 pKa = 8.37SATPPVLTISTLADD1717 pKa = 3.25GSVTNNATLNVSGQATSINGITSVTVNGAAVTLAADD1753 pKa = 4.04GTFSQAVTLQAGTNTVTTIATDD1775 pKa = 3.49NAGTTTTDD1783 pKa = 2.49SRR1785 pKa = 11.84TITLDD1790 pKa = 2.78TTAPVITITTPADD1803 pKa = 3.43NSTLAASSVTITGSVDD1819 pKa = 2.92KK1820 pKa = 10.79NATVQATANSGSPQSAAMTNNTFTVTLNLAGGSNTIVISATDD1862 pKa = 3.6LAGNSASVKK1871 pKa = 9.12RR1872 pKa = 11.84TIVSDD1877 pKa = 3.48TTKK1880 pKa = 9.41PTLAITNPSQDD1891 pKa = 2.88ITISEE1896 pKa = 4.25PALTISGTVTDD1907 pKa = 4.6ALTDD1911 pKa = 3.41VSVTITCDD1919 pKa = 2.83GKK1921 pKa = 9.18TYY1923 pKa = 10.05TPQVVDD1929 pKa = 4.49GAFQQQLTFVMAKK1942 pKa = 9.48QYY1944 pKa = 10.93AITVTATDD1952 pKa = 3.34QAGNSVTTQRR1962 pKa = 11.84NVIYY1966 pKa = 9.92ALSSLPSGDD1975 pKa = 3.15INGDD1979 pKa = 3.62GKK1981 pKa = 11.42VDD1983 pKa = 3.26IADD1986 pKa = 4.06ALLALQMAVGLITPTSAQLATGDD2009 pKa = 4.0VAPLSNGKK2017 pKa = 8.45PAPDD2021 pKa = 3.62GAIDD2025 pKa = 3.6IADD2028 pKa = 3.46AMLILEE2034 pKa = 4.79KK2035 pKa = 10.64AVGMLTWW2042 pKa = 4.01
Molecular weight: 199.61 kDa
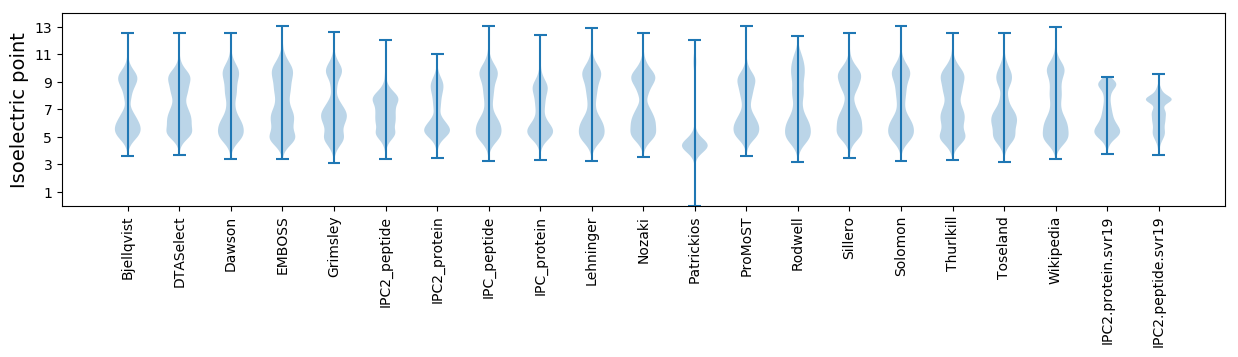
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A5G9W5|A5G9W5_GEOUR SAM-dependent methyltransferase OS=Geobacter uraniireducens (strain Rf4) OX=351605 GN=Gura_1435 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.33QPSNVSRR12 pKa = 11.84KK13 pKa = 7.52RR14 pKa = 11.84THH16 pKa = 6.03GFLVRR21 pKa = 11.84MSTKK25 pKa = 10.31NGRR28 pKa = 11.84LVIKK32 pKa = 10.39RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.01GRR39 pKa = 11.84KK40 pKa = 8.3NLAVSIASKK49 pKa = 10.89
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.33QPSNVSRR12 pKa = 11.84KK13 pKa = 7.52RR14 pKa = 11.84THH16 pKa = 6.03GFLVRR21 pKa = 11.84MSTKK25 pKa = 10.31NGRR28 pKa = 11.84LVIKK32 pKa = 10.39RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.01GRR39 pKa = 11.84KK40 pKa = 8.3NLAVSIASKK49 pKa = 10.89
Molecular weight: 5.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1447873 |
31 |
7157 |
340.3 |
37.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.292 ± 0.05 | 1.28 ± 0.026 |
5.252 ± 0.031 | 6.396 ± 0.06 |
4.11 ± 0.027 | 8.001 ± 0.044 |
1.993 ± 0.025 | 6.226 ± 0.034 |
5.416 ± 0.041 | 9.91 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.469 ± 0.019 | 3.738 ± 0.038 |
4.374 ± 0.028 | 3.041 ± 0.024 |
5.711 ± 0.046 | 5.953 ± 0.038 |
5.615 ± 0.065 | 7.225 ± 0.033 |
1.027 ± 0.016 | 2.97 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
