
Bombilactobacillus mellis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Bombilactobacillus
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
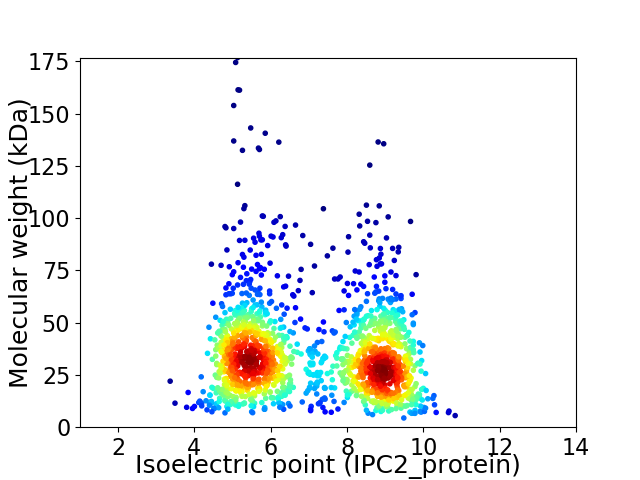
Virtual 2D-PAGE plot for 1572 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F4KQ95|A0A0F4KQ95_9LACO Putative transcriptional regulator lytR family OS=Bombilactobacillus mellis OX=1218508 GN=JG29_15200 PE=3 SV=1
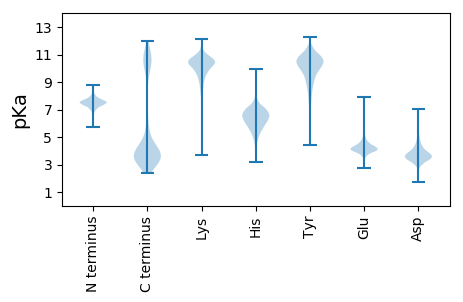
MM1 pKa = 7.23TKK3 pKa = 10.1IKK5 pKa = 10.13IVYY8 pKa = 10.51ASMTGNDD15 pKa = 3.63EE16 pKa = 4.95DD17 pKa = 3.99IADD20 pKa = 3.84ILSEE24 pKa = 4.2DD25 pKa = 3.82FEE27 pKa = 4.58EE28 pKa = 6.64AGMQVDD34 pKa = 4.64LSEE37 pKa = 4.6ISQTDD42 pKa = 3.47VADD45 pKa = 3.63YY46 pKa = 10.6LQADD50 pKa = 3.89VCIMVSYY57 pKa = 9.54TYY59 pKa = 11.24SDD61 pKa = 3.43GLEE64 pKa = 4.01GNIPDD69 pKa = 4.15EE70 pKa = 4.6GLDD73 pKa = 3.81FYY75 pKa = 11.61HH76 pKa = 7.71DD77 pKa = 4.33LLEE80 pKa = 5.56SDD82 pKa = 4.91LSNKK86 pKa = 9.34IFGVCGSGDD95 pKa = 3.15TFYY98 pKa = 10.95PEE100 pKa = 3.65FCVAVDD106 pKa = 3.57QFEE109 pKa = 4.38KK110 pKa = 11.19AFLQAHH116 pKa = 6.57ARR118 pKa = 11.84QGAKK122 pKa = 9.4SVKK125 pKa = 9.88IEE127 pKa = 4.05LAPNNDD133 pKa = 4.7DD134 pKa = 4.36IVKK137 pKa = 10.01LDD139 pKa = 4.21HH140 pKa = 6.55FAQTIVEE147 pKa = 4.2NCQQ150 pKa = 2.86
MM1 pKa = 7.23TKK3 pKa = 10.1IKK5 pKa = 10.13IVYY8 pKa = 10.51ASMTGNDD15 pKa = 3.63EE16 pKa = 4.95DD17 pKa = 3.99IADD20 pKa = 3.84ILSEE24 pKa = 4.2DD25 pKa = 3.82FEE27 pKa = 4.58EE28 pKa = 6.64AGMQVDD34 pKa = 4.64LSEE37 pKa = 4.6ISQTDD42 pKa = 3.47VADD45 pKa = 3.63YY46 pKa = 10.6LQADD50 pKa = 3.89VCIMVSYY57 pKa = 9.54TYY59 pKa = 11.24SDD61 pKa = 3.43GLEE64 pKa = 4.01GNIPDD69 pKa = 4.15EE70 pKa = 4.6GLDD73 pKa = 3.81FYY75 pKa = 11.61HH76 pKa = 7.71DD77 pKa = 4.33LLEE80 pKa = 5.56SDD82 pKa = 4.91LSNKK86 pKa = 9.34IFGVCGSGDD95 pKa = 3.15TFYY98 pKa = 10.95PEE100 pKa = 3.65FCVAVDD106 pKa = 3.57QFEE109 pKa = 4.38KK110 pKa = 11.19AFLQAHH116 pKa = 6.57ARR118 pKa = 11.84QGAKK122 pKa = 9.4SVKK125 pKa = 9.88IEE127 pKa = 4.05LAPNNDD133 pKa = 4.7DD134 pKa = 4.36IVKK137 pKa = 10.01LDD139 pKa = 4.21HH140 pKa = 6.55FAQTIVEE147 pKa = 4.2NCQQ150 pKa = 2.86
Molecular weight: 16.62 kDa
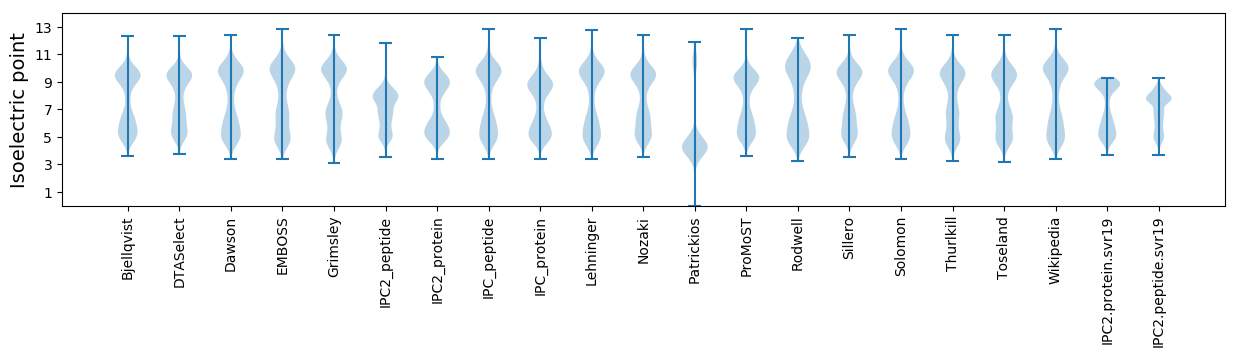
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F4KYU7|A0A0F4KYU7_9LACO L-ribulose-5-phosphate 4-epimerase OS=Bombilactobacillus mellis OX=1218508 GN=JG29_01680 PE=4 SV=1
MM1 pKa = 6.82TKK3 pKa = 10.26KK4 pKa = 9.84RR5 pKa = 11.84LKK7 pKa = 10.19VISIILIIVAFLLVFAGYY25 pKa = 9.39RR26 pKa = 11.84QQTAALVPTRR36 pKa = 11.84LHH38 pKa = 5.3STPTLFLHH46 pKa = 6.8GWGSSINAEE55 pKa = 4.05KK56 pKa = 10.87QMTSAAQRR64 pKa = 11.84AGVTNSVTQALVDD77 pKa = 3.63RR78 pKa = 11.84QGRR81 pKa = 11.84VHH83 pKa = 7.14LKK85 pKa = 10.22GHH87 pKa = 6.84IPPHH91 pKa = 5.68ARR93 pKa = 11.84NPIIEE98 pKa = 4.34VGFKK102 pKa = 10.8DD103 pKa = 3.63NRR105 pKa = 11.84NPNYY109 pKa = 10.15HH110 pKa = 6.6QDD112 pKa = 3.69GRR114 pKa = 11.84WLKK117 pKa = 10.32QVILKK122 pKa = 9.03LQQQYY127 pKa = 10.03QIKK130 pKa = 9.66NINLVGHH137 pKa = 6.07SMGNMAISYY146 pKa = 9.71YY147 pKa = 11.14LLDD150 pKa = 3.84NADD153 pKa = 3.6NQRR156 pKa = 11.84LPKK159 pKa = 10.03LRR161 pKa = 11.84KK162 pKa = 7.8QVAIAGHH169 pKa = 5.63FAGIIGAGDD178 pKa = 3.45RR179 pKa = 11.84PHH181 pKa = 7.27RR182 pKa = 11.84LHH184 pKa = 6.97LAATGRR190 pKa = 11.84PNHH193 pKa = 6.35MDD195 pKa = 3.07NNYY198 pKa = 7.25RR199 pKa = 11.84TLMGLRR205 pKa = 11.84QRR207 pKa = 11.84YY208 pKa = 7.93PNKK211 pKa = 9.46QVKK214 pKa = 9.05VLNIFGDD221 pKa = 4.0KK222 pKa = 10.89NDD224 pKa = 4.05GTNSDD229 pKa = 3.7GDD231 pKa = 3.86VTNASAQSLRR241 pKa = 11.84YY242 pKa = 9.15LVQARR247 pKa = 11.84ALSYY251 pKa = 9.09QEE253 pKa = 3.99RR254 pKa = 11.84KK255 pKa = 9.63IVGSDD260 pKa = 3.41GQHH263 pKa = 5.44SRR265 pKa = 11.84LHH267 pKa = 5.44EE268 pKa = 4.17SRR270 pKa = 11.84RR271 pKa = 11.84VDD273 pKa = 3.76RR274 pKa = 11.84ILINFLWKK282 pKa = 10.52
MM1 pKa = 6.82TKK3 pKa = 10.26KK4 pKa = 9.84RR5 pKa = 11.84LKK7 pKa = 10.19VISIILIIVAFLLVFAGYY25 pKa = 9.39RR26 pKa = 11.84QQTAALVPTRR36 pKa = 11.84LHH38 pKa = 5.3STPTLFLHH46 pKa = 6.8GWGSSINAEE55 pKa = 4.05KK56 pKa = 10.87QMTSAAQRR64 pKa = 11.84AGVTNSVTQALVDD77 pKa = 3.63RR78 pKa = 11.84QGRR81 pKa = 11.84VHH83 pKa = 7.14LKK85 pKa = 10.22GHH87 pKa = 6.84IPPHH91 pKa = 5.68ARR93 pKa = 11.84NPIIEE98 pKa = 4.34VGFKK102 pKa = 10.8DD103 pKa = 3.63NRR105 pKa = 11.84NPNYY109 pKa = 10.15HH110 pKa = 6.6QDD112 pKa = 3.69GRR114 pKa = 11.84WLKK117 pKa = 10.32QVILKK122 pKa = 9.03LQQQYY127 pKa = 10.03QIKK130 pKa = 9.66NINLVGHH137 pKa = 6.07SMGNMAISYY146 pKa = 9.71YY147 pKa = 11.14LLDD150 pKa = 3.84NADD153 pKa = 3.6NQRR156 pKa = 11.84LPKK159 pKa = 10.03LRR161 pKa = 11.84KK162 pKa = 7.8QVAIAGHH169 pKa = 5.63FAGIIGAGDD178 pKa = 3.45RR179 pKa = 11.84PHH181 pKa = 7.27RR182 pKa = 11.84LHH184 pKa = 6.97LAATGRR190 pKa = 11.84PNHH193 pKa = 6.35MDD195 pKa = 3.07NNYY198 pKa = 7.25RR199 pKa = 11.84TLMGLRR205 pKa = 11.84QRR207 pKa = 11.84YY208 pKa = 7.93PNKK211 pKa = 9.46QVKK214 pKa = 9.05VLNIFGDD221 pKa = 4.0KK222 pKa = 10.89NDD224 pKa = 4.05GTNSDD229 pKa = 3.7GDD231 pKa = 3.86VTNASAQSLRR241 pKa = 11.84YY242 pKa = 9.15LVQARR247 pKa = 11.84ALSYY251 pKa = 9.09QEE253 pKa = 3.99RR254 pKa = 11.84KK255 pKa = 9.63IVGSDD260 pKa = 3.41GQHH263 pKa = 5.44SRR265 pKa = 11.84LHH267 pKa = 5.44EE268 pKa = 4.17SRR270 pKa = 11.84RR271 pKa = 11.84VDD273 pKa = 3.76RR274 pKa = 11.84ILINFLWKK282 pKa = 10.52
Molecular weight: 31.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
510670 |
38 |
1594 |
324.9 |
36.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.681 ± 0.066 | 0.622 ± 0.015 |
5.172 ± 0.053 | 4.568 ± 0.059 |
4.253 ± 0.047 | 6.146 ± 0.06 |
2.258 ± 0.028 | 7.824 ± 0.057 |
6.322 ± 0.053 | 10.446 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.338 ± 0.031 | 5.49 ± 0.072 |
3.771 ± 0.035 | 6.505 ± 0.082 |
3.652 ± 0.048 | 5.886 ± 0.056 |
5.8 ± 0.054 | 6.448 ± 0.05 |
1.084 ± 0.023 | 3.735 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
