
Tenacibaculum sp. SZ-18
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum; unclassified Tenacibaculum
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
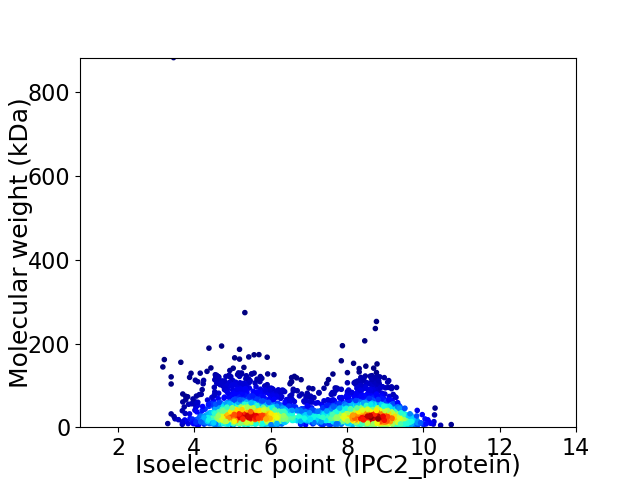
Virtual 2D-PAGE plot for 3514 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K8VDC7|A0A2K8VDC7_9FLAO ANK_REP_REGION domain-containing protein OS=Tenacibaculum sp. SZ-18 OX=754423 GN=BTO06_11215 PE=4 SV=1
MM1 pKa = 7.69LSLNIYY7 pKa = 9.06GQTIDD12 pKa = 4.14KK13 pKa = 9.67PILGFSSACGSDD25 pKa = 2.86GFNNFNFSFVFRR37 pKa = 11.84DD38 pKa = 3.82GLFDD42 pKa = 3.79TNNTFNVVISDD53 pKa = 3.66EE54 pKa = 4.4NGDD57 pKa = 3.75FGANPPVIFSITDD70 pKa = 3.38SNVNTFPTNPVDD82 pKa = 4.08GSFSVPDD89 pKa = 3.89DD90 pKa = 3.86AFGTSYY96 pKa = 10.53RR97 pKa = 11.84IKK99 pKa = 10.8VIANSPNIEE108 pKa = 4.57SPPSDD113 pKa = 3.8PFEE116 pKa = 4.14MYY118 pKa = 10.26RR119 pKa = 11.84ISSEE123 pKa = 3.85PLILNEE129 pKa = 4.44FNDD132 pKa = 5.44DD133 pKa = 3.81LVLCNGVAEE142 pKa = 4.43TVSLNLTDD150 pKa = 4.69PNVEE154 pKa = 4.6FIWYY158 pKa = 9.6RR159 pKa = 11.84NNTPITSQSSSSLTITQPGEE179 pKa = 3.88YY180 pKa = 9.28YY181 pKa = 11.11AEE183 pKa = 4.18INYY186 pKa = 9.13GGCPDD191 pKa = 3.97AVISNKK197 pKa = 10.0VIARR201 pKa = 11.84VVNSSSITIQGDD213 pKa = 3.52NTVEE217 pKa = 3.96LCADD221 pKa = 3.34QAYY224 pKa = 10.76DD225 pKa = 4.87LIASIDD231 pKa = 4.02DD232 pKa = 3.57PSFIYY237 pKa = 10.02KK238 pKa = 9.1WFKK241 pKa = 10.76DD242 pKa = 3.49GNEE245 pKa = 3.86ITGLPDD251 pKa = 3.04YY252 pKa = 10.31TPIYY256 pKa = 6.89TTPTSDD262 pKa = 3.01QFGVYY267 pKa = 9.6HH268 pKa = 6.71LEE270 pKa = 4.1IEE272 pKa = 4.27ISGCSSRR279 pKa = 11.84SQDD282 pKa = 2.95VTLEE286 pKa = 4.12QKK288 pKa = 10.57SDD290 pKa = 2.88AGFPVEE296 pKa = 4.31ISGNPTEE303 pKa = 4.56IVFPSEE309 pKa = 4.09EE310 pKa = 3.89FTLTVTHH317 pKa = 6.42EE318 pKa = 3.98ASGPVDD324 pKa = 3.51IKK326 pKa = 10.84WFDD329 pKa = 3.9ANGEE333 pKa = 4.28IPGATGDD340 pKa = 3.91TLNVSGPGEE349 pKa = 4.22YY350 pKa = 9.92YY351 pKa = 11.1AEE353 pKa = 4.24VTEE356 pKa = 4.86SGGDD360 pKa = 3.67CPVSVLSDD368 pKa = 2.82IRR370 pKa = 11.84EE371 pKa = 3.9ILGVKK376 pKa = 9.95EE377 pKa = 3.82MFVTIRR383 pKa = 11.84SGTDD387 pKa = 2.96YY388 pKa = 11.01DD389 pKa = 3.98EE390 pKa = 5.11CVSASTEE397 pKa = 3.83LTMVGISVIATDD409 pKa = 3.84DD410 pKa = 3.54NEE412 pKa = 4.42YY413 pKa = 11.07DD414 pKa = 3.61VSQEE418 pKa = 4.61KK419 pKa = 9.71IDD421 pKa = 4.25KK422 pKa = 10.31LLSPDD427 pKa = 3.16STGAIIMRR435 pKa = 11.84FQWYY439 pKa = 10.33KK440 pKa = 10.94NGTTITDD447 pKa = 3.68AEE449 pKa = 4.57SILYY453 pKa = 9.21NVNSYY458 pKa = 11.36AEE460 pKa = 3.88NDD462 pKa = 3.38EE463 pKa = 4.55YY464 pKa = 11.69YY465 pKa = 11.27LNLEE469 pKa = 4.31VGSLSADD476 pKa = 3.64SNILDD481 pKa = 3.42ILLGVGEE488 pKa = 4.2VQIVSSSVSNALCPGEE504 pKa = 5.21SIFLSITDD512 pKa = 3.76FTGYY516 pKa = 10.38SYY518 pKa = 10.38TWFKK522 pKa = 11.31DD523 pKa = 3.75GIPITVSDD531 pKa = 4.33PLNIEE536 pKa = 3.71VSEE539 pKa = 4.08IGSYY543 pKa = 10.85SVTLDD548 pKa = 4.13GFGCQINIAEE558 pKa = 4.16VEE560 pKa = 4.09IVEE563 pKa = 4.61FDD565 pKa = 3.65DD566 pKa = 3.87TVLEE570 pKa = 4.31VSPSTNAVLPVGGSVTFEE588 pKa = 3.64ASGADD593 pKa = 3.23SYY595 pKa = 11.59EE596 pKa = 4.15WYY598 pKa = 10.25DD599 pKa = 3.37EE600 pKa = 4.27AGNLLSTNEE609 pKa = 4.04TLEE612 pKa = 4.07VNTLGTYY619 pKa = 9.85IVIGTVGGCTAQRR632 pKa = 11.84EE633 pKa = 4.63VNVVEE638 pKa = 5.23DD639 pKa = 4.29DD640 pKa = 4.11GKK642 pKa = 11.32LVIPNIITPFNGDD655 pKa = 3.93GINDD659 pKa = 3.16KK660 pKa = 10.73WEE662 pKa = 4.22LPNRR666 pKa = 11.84FAFQTNVQILIFNSKK681 pKa = 10.58GEE683 pKa = 4.22EE684 pKa = 4.02VLNTTDD690 pKa = 3.91YY691 pKa = 10.95QNNWPEE697 pKa = 4.18DD698 pKa = 3.52NNLRR702 pKa = 11.84DD703 pKa = 3.36GMLFYY708 pKa = 10.77FKK710 pKa = 10.67VIRR713 pKa = 11.84EE714 pKa = 4.06NNLIKK719 pKa = 10.77AGTISILQQ727 pKa = 3.5
MM1 pKa = 7.69LSLNIYY7 pKa = 9.06GQTIDD12 pKa = 4.14KK13 pKa = 9.67PILGFSSACGSDD25 pKa = 2.86GFNNFNFSFVFRR37 pKa = 11.84DD38 pKa = 3.82GLFDD42 pKa = 3.79TNNTFNVVISDD53 pKa = 3.66EE54 pKa = 4.4NGDD57 pKa = 3.75FGANPPVIFSITDD70 pKa = 3.38SNVNTFPTNPVDD82 pKa = 4.08GSFSVPDD89 pKa = 3.89DD90 pKa = 3.86AFGTSYY96 pKa = 10.53RR97 pKa = 11.84IKK99 pKa = 10.8VIANSPNIEE108 pKa = 4.57SPPSDD113 pKa = 3.8PFEE116 pKa = 4.14MYY118 pKa = 10.26RR119 pKa = 11.84ISSEE123 pKa = 3.85PLILNEE129 pKa = 4.44FNDD132 pKa = 5.44DD133 pKa = 3.81LVLCNGVAEE142 pKa = 4.43TVSLNLTDD150 pKa = 4.69PNVEE154 pKa = 4.6FIWYY158 pKa = 9.6RR159 pKa = 11.84NNTPITSQSSSSLTITQPGEE179 pKa = 3.88YY180 pKa = 9.28YY181 pKa = 11.11AEE183 pKa = 4.18INYY186 pKa = 9.13GGCPDD191 pKa = 3.97AVISNKK197 pKa = 10.0VIARR201 pKa = 11.84VVNSSSITIQGDD213 pKa = 3.52NTVEE217 pKa = 3.96LCADD221 pKa = 3.34QAYY224 pKa = 10.76DD225 pKa = 4.87LIASIDD231 pKa = 4.02DD232 pKa = 3.57PSFIYY237 pKa = 10.02KK238 pKa = 9.1WFKK241 pKa = 10.76DD242 pKa = 3.49GNEE245 pKa = 3.86ITGLPDD251 pKa = 3.04YY252 pKa = 10.31TPIYY256 pKa = 6.89TTPTSDD262 pKa = 3.01QFGVYY267 pKa = 9.6HH268 pKa = 6.71LEE270 pKa = 4.1IEE272 pKa = 4.27ISGCSSRR279 pKa = 11.84SQDD282 pKa = 2.95VTLEE286 pKa = 4.12QKK288 pKa = 10.57SDD290 pKa = 2.88AGFPVEE296 pKa = 4.31ISGNPTEE303 pKa = 4.56IVFPSEE309 pKa = 4.09EE310 pKa = 3.89FTLTVTHH317 pKa = 6.42EE318 pKa = 3.98ASGPVDD324 pKa = 3.51IKK326 pKa = 10.84WFDD329 pKa = 3.9ANGEE333 pKa = 4.28IPGATGDD340 pKa = 3.91TLNVSGPGEE349 pKa = 4.22YY350 pKa = 9.92YY351 pKa = 11.1AEE353 pKa = 4.24VTEE356 pKa = 4.86SGGDD360 pKa = 3.67CPVSVLSDD368 pKa = 2.82IRR370 pKa = 11.84EE371 pKa = 3.9ILGVKK376 pKa = 9.95EE377 pKa = 3.82MFVTIRR383 pKa = 11.84SGTDD387 pKa = 2.96YY388 pKa = 11.01DD389 pKa = 3.98EE390 pKa = 5.11CVSASTEE397 pKa = 3.83LTMVGISVIATDD409 pKa = 3.84DD410 pKa = 3.54NEE412 pKa = 4.42YY413 pKa = 11.07DD414 pKa = 3.61VSQEE418 pKa = 4.61KK419 pKa = 9.71IDD421 pKa = 4.25KK422 pKa = 10.31LLSPDD427 pKa = 3.16STGAIIMRR435 pKa = 11.84FQWYY439 pKa = 10.33KK440 pKa = 10.94NGTTITDD447 pKa = 3.68AEE449 pKa = 4.57SILYY453 pKa = 9.21NVNSYY458 pKa = 11.36AEE460 pKa = 3.88NDD462 pKa = 3.38EE463 pKa = 4.55YY464 pKa = 11.69YY465 pKa = 11.27LNLEE469 pKa = 4.31VGSLSADD476 pKa = 3.64SNILDD481 pKa = 3.42ILLGVGEE488 pKa = 4.2VQIVSSSVSNALCPGEE504 pKa = 5.21SIFLSITDD512 pKa = 3.76FTGYY516 pKa = 10.38SYY518 pKa = 10.38TWFKK522 pKa = 11.31DD523 pKa = 3.75GIPITVSDD531 pKa = 4.33PLNIEE536 pKa = 3.71VSEE539 pKa = 4.08IGSYY543 pKa = 10.85SVTLDD548 pKa = 4.13GFGCQINIAEE558 pKa = 4.16VEE560 pKa = 4.09IVEE563 pKa = 4.61FDD565 pKa = 3.65DD566 pKa = 3.87TVLEE570 pKa = 4.31VSPSTNAVLPVGGSVTFEE588 pKa = 3.64ASGADD593 pKa = 3.23SYY595 pKa = 11.59EE596 pKa = 4.15WYY598 pKa = 10.25DD599 pKa = 3.37EE600 pKa = 4.27AGNLLSTNEE609 pKa = 4.04TLEE612 pKa = 4.07VNTLGTYY619 pKa = 9.85IVIGTVGGCTAQRR632 pKa = 11.84EE633 pKa = 4.63VNVVEE638 pKa = 5.23DD639 pKa = 4.29DD640 pKa = 4.11GKK642 pKa = 11.32LVIPNIITPFNGDD655 pKa = 3.93GINDD659 pKa = 3.16KK660 pKa = 10.73WEE662 pKa = 4.22LPNRR666 pKa = 11.84FAFQTNVQILIFNSKK681 pKa = 10.58GEE683 pKa = 4.22EE684 pKa = 4.02VLNTTDD690 pKa = 3.91YY691 pKa = 10.95QNNWPEE697 pKa = 4.18DD698 pKa = 3.52NNLRR702 pKa = 11.84DD703 pKa = 3.36GMLFYY708 pKa = 10.77FKK710 pKa = 10.67VIRR713 pKa = 11.84EE714 pKa = 4.06NNLIKK719 pKa = 10.77AGTISILQQ727 pKa = 3.5
Molecular weight: 79.31 kDa
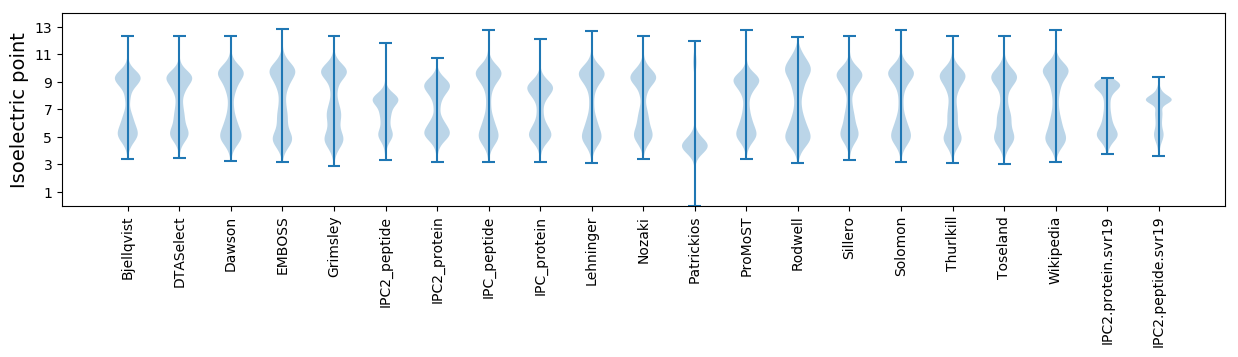
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K8V9F7|A0A2K8V9F7_9FLAO Histidine kinase OS=Tenacibaculum sp. SZ-18 OX=754423 GN=BTO06_03230 PE=4 SV=1
MM1 pKa = 7.6INGIRR6 pKa = 11.84YY7 pKa = 7.92FFEE10 pKa = 5.13RR11 pKa = 11.84NGFDD15 pKa = 3.1VSSRR19 pKa = 11.84LADD22 pKa = 3.81RR23 pKa = 11.84LGMRR27 pKa = 11.84AVNVRR32 pKa = 11.84LFFIYY37 pKa = 10.25ISFVTVGLSFALYY50 pKa = 10.19LVIAFLLRR58 pKa = 11.84LKK60 pKa = 10.86DD61 pKa = 3.81RR62 pKa = 11.84IYY64 pKa = 10.89TKK66 pKa = 9.75RR67 pKa = 11.84TSVFDD72 pKa = 3.66LL73 pKa = 4.18
MM1 pKa = 7.6INGIRR6 pKa = 11.84YY7 pKa = 7.92FFEE10 pKa = 5.13RR11 pKa = 11.84NGFDD15 pKa = 3.1VSSRR19 pKa = 11.84LADD22 pKa = 3.81RR23 pKa = 11.84LGMRR27 pKa = 11.84AVNVRR32 pKa = 11.84LFFIYY37 pKa = 10.25ISFVTVGLSFALYY50 pKa = 10.19LVIAFLLRR58 pKa = 11.84LKK60 pKa = 10.86DD61 pKa = 3.81RR62 pKa = 11.84IYY64 pKa = 10.89TKK66 pKa = 9.75RR67 pKa = 11.84TSVFDD72 pKa = 3.66LL73 pKa = 4.18
Molecular weight: 8.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1157436 |
30 |
8531 |
329.4 |
37.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.429 ± 0.043 | 0.727 ± 0.017 |
5.441 ± 0.032 | 6.751 ± 0.049 |
5.413 ± 0.035 | 6.08 ± 0.059 |
1.649 ± 0.022 | 8.345 ± 0.045 |
8.605 ± 0.074 | 9.203 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.954 ± 0.022 | 6.824 ± 0.038 |
3.087 ± 0.028 | 3.23 ± 0.022 |
3.423 ± 0.029 | 6.864 ± 0.043 |
5.744 ± 0.077 | 6.05 ± 0.039 |
1.011 ± 0.015 | 4.171 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
