
bacterium 0.1xD8-82
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
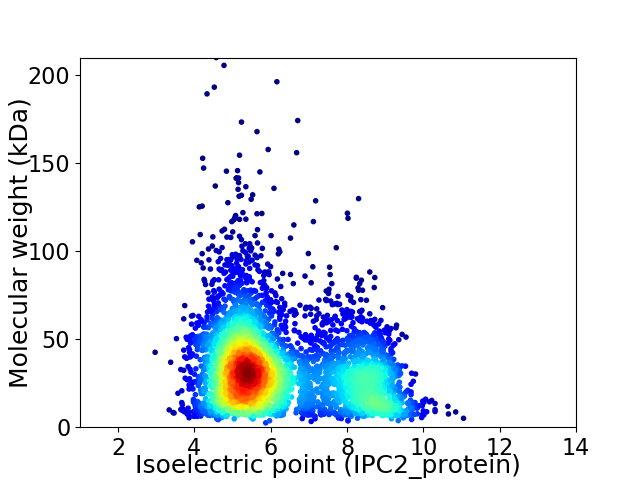
Virtual 2D-PAGE plot for 4335 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A9ATI9|A0A3A9ATI9_9BACT Uncharacterized protein OS=bacterium 0.1xD8-82 OX=2320100 GN=D7V94_12135 PE=4 SV=1
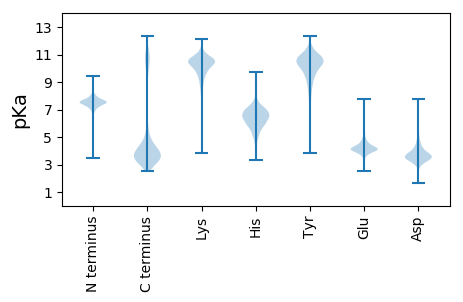
MM1 pKa = 7.21KK2 pKa = 10.16RR3 pKa = 11.84KK4 pKa = 9.29IVAVLLSAAMILSMTACGNSSSGSNASEE32 pKa = 4.26GAAQDD37 pKa = 3.52TAQAADD43 pKa = 3.82TSNEE47 pKa = 3.76DD48 pKa = 3.39AAADD52 pKa = 3.81AQDD55 pKa = 4.54ADD57 pKa = 4.57AEE59 pKa = 4.26DD60 pKa = 4.19SADD63 pKa = 3.61EE64 pKa = 4.55GASSSATADD73 pKa = 3.57FSGVKK78 pKa = 10.34AEE80 pKa = 4.1IDD82 pKa = 3.01IDD84 pKa = 3.78GTIIGDD90 pKa = 3.64ALTTFEE96 pKa = 4.65GKK98 pKa = 10.0VEE100 pKa = 4.06EE101 pKa = 4.47FNNLTGAQLEE111 pKa = 4.36LVEE114 pKa = 5.47NGDD117 pKa = 3.61DD118 pKa = 3.94HH119 pKa = 6.41EE120 pKa = 7.63AIMKK124 pKa = 8.9TRR126 pKa = 11.84MASNDD131 pKa = 3.73MPDD134 pKa = 2.73MFTTHH139 pKa = 5.21GWSTIRR145 pKa = 11.84YY146 pKa = 9.22NEE148 pKa = 3.71FCYY151 pKa = 10.72DD152 pKa = 3.8LSGEE156 pKa = 4.25SWVSDD161 pKa = 3.54MADD164 pKa = 2.84AAAAVVTDD172 pKa = 4.6PNGKK176 pKa = 8.85ICTCPLTEE184 pKa = 3.97WVYY187 pKa = 11.2GIAFNNTIMEE197 pKa = 4.79DD198 pKa = 3.41NGIDD202 pKa = 3.28MYY204 pKa = 11.3AIKK207 pKa = 9.08TWDD210 pKa = 4.14DD211 pKa = 3.28LTAALQTLKK220 pKa = 10.86DD221 pKa = 3.65AGITPISVGGKK232 pKa = 7.85STGSLGGYY240 pKa = 10.18LEE242 pKa = 4.31MSNVFYY248 pKa = 11.06SADD251 pKa = 2.97GAMYY255 pKa = 10.58DD256 pKa = 5.19GGAQLQDD263 pKa = 3.06GSFSFVEE270 pKa = 4.14HH271 pKa = 6.89PEE273 pKa = 4.21IIARR277 pKa = 11.84FADD280 pKa = 4.44LYY282 pKa = 11.33DD283 pKa = 3.41KK284 pKa = 11.54GFFIEE289 pKa = 5.57DD290 pKa = 4.61LFTADD295 pKa = 4.11PDD297 pKa = 3.84TARR300 pKa = 11.84NYY302 pKa = 10.15VATGQAAMLLWGSPEE317 pKa = 3.92YY318 pKa = 10.9VDD320 pKa = 5.59LMKK323 pKa = 10.14TANPDD328 pKa = 3.31NEE330 pKa = 4.27FGIIPVPAVEE340 pKa = 4.61EE341 pKa = 4.5GGKK344 pKa = 8.84PAYY347 pKa = 9.16TVGEE351 pKa = 4.43GTAIAISSSTEE362 pKa = 3.7NLEE365 pKa = 3.99LCQAFLNFLTDD376 pKa = 3.6PEE378 pKa = 4.4TLKK381 pKa = 11.09VYY383 pKa = 10.63VDD385 pKa = 3.36STGAVSGFKK394 pKa = 10.03TISQDD399 pKa = 3.5DD400 pKa = 4.41TYY402 pKa = 11.93SLNKK406 pKa = 10.0YY407 pKa = 9.56NEE409 pKa = 4.6SIEE412 pKa = 4.06KK413 pKa = 9.44VANITYY419 pKa = 9.87TNFFDD424 pKa = 5.57RR425 pKa = 11.84EE426 pKa = 4.14YY427 pKa = 10.63LPSGMWNYY435 pKa = 9.37MSEE438 pKa = 4.78SIAQLFNCNVGEE450 pKa = 4.2ASGKK454 pKa = 8.5VDD456 pKa = 3.65EE457 pKa = 4.66VAEE460 pKa = 4.14YY461 pKa = 8.65MQQAYY466 pKa = 8.27EE467 pKa = 4.03QLYY470 pKa = 8.42ATNNEE475 pKa = 3.98
MM1 pKa = 7.21KK2 pKa = 10.16RR3 pKa = 11.84KK4 pKa = 9.29IVAVLLSAAMILSMTACGNSSSGSNASEE32 pKa = 4.26GAAQDD37 pKa = 3.52TAQAADD43 pKa = 3.82TSNEE47 pKa = 3.76DD48 pKa = 3.39AAADD52 pKa = 3.81AQDD55 pKa = 4.54ADD57 pKa = 4.57AEE59 pKa = 4.26DD60 pKa = 4.19SADD63 pKa = 3.61EE64 pKa = 4.55GASSSATADD73 pKa = 3.57FSGVKK78 pKa = 10.34AEE80 pKa = 4.1IDD82 pKa = 3.01IDD84 pKa = 3.78GTIIGDD90 pKa = 3.64ALTTFEE96 pKa = 4.65GKK98 pKa = 10.0VEE100 pKa = 4.06EE101 pKa = 4.47FNNLTGAQLEE111 pKa = 4.36LVEE114 pKa = 5.47NGDD117 pKa = 3.61DD118 pKa = 3.94HH119 pKa = 6.41EE120 pKa = 7.63AIMKK124 pKa = 8.9TRR126 pKa = 11.84MASNDD131 pKa = 3.73MPDD134 pKa = 2.73MFTTHH139 pKa = 5.21GWSTIRR145 pKa = 11.84YY146 pKa = 9.22NEE148 pKa = 3.71FCYY151 pKa = 10.72DD152 pKa = 3.8LSGEE156 pKa = 4.25SWVSDD161 pKa = 3.54MADD164 pKa = 2.84AAAAVVTDD172 pKa = 4.6PNGKK176 pKa = 8.85ICTCPLTEE184 pKa = 3.97WVYY187 pKa = 11.2GIAFNNTIMEE197 pKa = 4.79DD198 pKa = 3.41NGIDD202 pKa = 3.28MYY204 pKa = 11.3AIKK207 pKa = 9.08TWDD210 pKa = 4.14DD211 pKa = 3.28LTAALQTLKK220 pKa = 10.86DD221 pKa = 3.65AGITPISVGGKK232 pKa = 7.85STGSLGGYY240 pKa = 10.18LEE242 pKa = 4.31MSNVFYY248 pKa = 11.06SADD251 pKa = 2.97GAMYY255 pKa = 10.58DD256 pKa = 5.19GGAQLQDD263 pKa = 3.06GSFSFVEE270 pKa = 4.14HH271 pKa = 6.89PEE273 pKa = 4.21IIARR277 pKa = 11.84FADD280 pKa = 4.44LYY282 pKa = 11.33DD283 pKa = 3.41KK284 pKa = 11.54GFFIEE289 pKa = 5.57DD290 pKa = 4.61LFTADD295 pKa = 4.11PDD297 pKa = 3.84TARR300 pKa = 11.84NYY302 pKa = 10.15VATGQAAMLLWGSPEE317 pKa = 3.92YY318 pKa = 10.9VDD320 pKa = 5.59LMKK323 pKa = 10.14TANPDD328 pKa = 3.31NEE330 pKa = 4.27FGIIPVPAVEE340 pKa = 4.61EE341 pKa = 4.5GGKK344 pKa = 8.84PAYY347 pKa = 9.16TVGEE351 pKa = 4.43GTAIAISSSTEE362 pKa = 3.7NLEE365 pKa = 3.99LCQAFLNFLTDD376 pKa = 3.6PEE378 pKa = 4.4TLKK381 pKa = 11.09VYY383 pKa = 10.63VDD385 pKa = 3.36STGAVSGFKK394 pKa = 10.03TISQDD399 pKa = 3.5DD400 pKa = 4.41TYY402 pKa = 11.93SLNKK406 pKa = 10.0YY407 pKa = 9.56NEE409 pKa = 4.6SIEE412 pKa = 4.06KK413 pKa = 9.44VANITYY419 pKa = 9.87TNFFDD424 pKa = 5.57RR425 pKa = 11.84EE426 pKa = 4.14YY427 pKa = 10.63LPSGMWNYY435 pKa = 9.37MSEE438 pKa = 4.78SIAQLFNCNVGEE450 pKa = 4.2ASGKK454 pKa = 8.5VDD456 pKa = 3.65EE457 pKa = 4.66VAEE460 pKa = 4.14YY461 pKa = 8.65MQQAYY466 pKa = 8.27EE467 pKa = 4.03QLYY470 pKa = 8.42ATNNEE475 pKa = 3.98
Molecular weight: 51.07 kDa
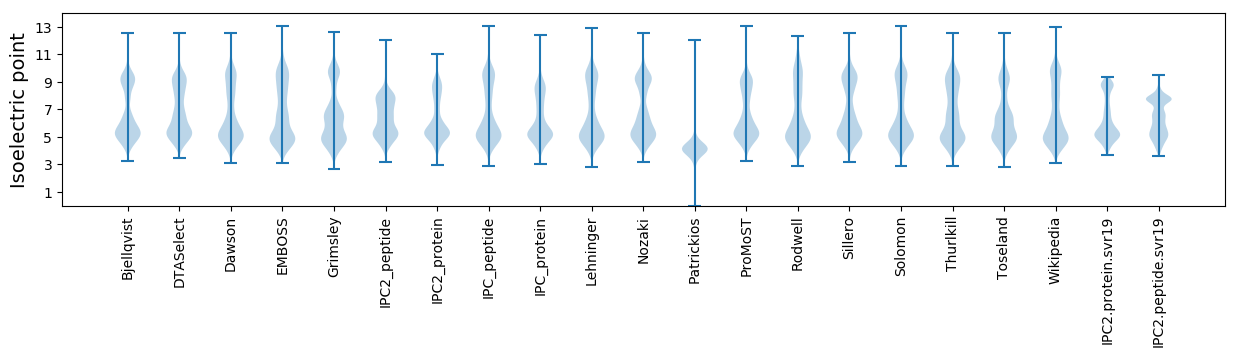
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9AWV5|A0A3A9AWV5_9BACT RNase H domain-containing protein OS=bacterium 0.1xD8-82 OX=2320100 GN=D7V94_08265 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1345899 |
25 |
1999 |
310.5 |
34.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.337 ± 0.037 | 1.597 ± 0.016 |
5.447 ± 0.033 | 7.721 ± 0.046 |
4.363 ± 0.031 | 7.171 ± 0.031 |
1.7 ± 0.016 | 7.364 ± 0.035 |
6.807 ± 0.034 | 9.114 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.176 ± 0.021 | 4.346 ± 0.028 |
3.252 ± 0.023 | 3.221 ± 0.02 |
4.659 ± 0.026 | 5.785 ± 0.032 |
4.986 ± 0.03 | 6.572 ± 0.031 |
1.031 ± 0.015 | 4.351 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
