
Fire ant associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses; Cruciviridae; Crucivirus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
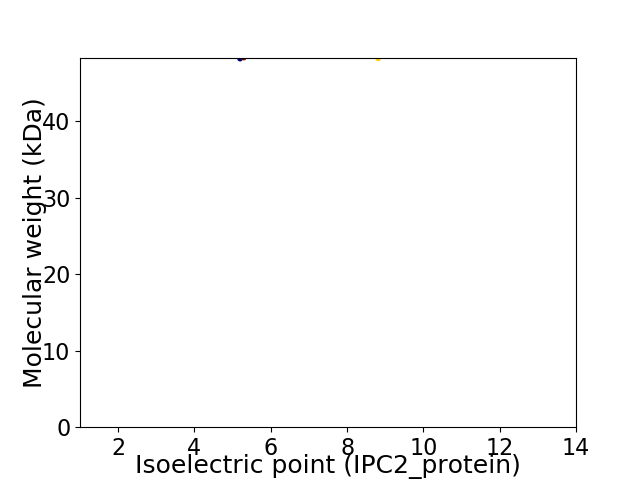
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPB1|A0A346BPB1_9VIRU Putative capsid protein OS=Fire ant associated circular virus 1 OX=2293280 PE=3 SV=1
MM1 pKa = 7.79ASTKK5 pKa = 10.25KK6 pKa = 10.46FRR8 pKa = 11.84FQNTSALLTYY18 pKa = 8.3KK19 pKa = 9.25THH21 pKa = 7.07LDD23 pKa = 3.04KK24 pKa = 10.96KK25 pKa = 10.89AYY27 pKa = 10.47LEE29 pKa = 4.12WFEE32 pKa = 4.61KK33 pKa = 9.95QAKK36 pKa = 5.67QTPVFIRR43 pKa = 11.84LAHH46 pKa = 5.47EE47 pKa = 4.52TGSHH51 pKa = 7.43DD52 pKa = 4.45EE53 pKa = 4.61GEE55 pKa = 4.05PDD57 pKa = 3.4YY58 pKa = 11.73DD59 pKa = 3.53HH60 pKa = 6.32THH62 pKa = 5.45VVFKK66 pKa = 10.48CKK68 pKa = 10.39SRR70 pKa = 11.84IDD72 pKa = 3.52TTSQRR77 pKa = 11.84FFDD80 pKa = 4.72FNAIHH85 pKa = 6.06PHH87 pKa = 5.17IKK89 pKa = 9.9VLKK92 pKa = 7.7TNKK95 pKa = 10.46SIDD98 pKa = 3.58DD99 pKa = 3.67AKK101 pKa = 11.08KK102 pKa = 10.78YY103 pKa = 9.13IAKK106 pKa = 9.67EE107 pKa = 4.0DD108 pKa = 3.86KK109 pKa = 10.91DD110 pKa = 4.45NEE112 pKa = 4.26DD113 pKa = 4.42LLVSDD118 pKa = 4.32TSNFIEE124 pKa = 5.0GVWSKK129 pKa = 9.28NTIQEE134 pKa = 4.06ALLDD138 pKa = 4.23FVKK141 pKa = 10.66KK142 pKa = 10.08PSDD145 pKa = 3.38VAGIIQLYY153 pKa = 8.88GLKK156 pKa = 10.41KK157 pKa = 10.56NPTQIHH163 pKa = 6.1EE164 pKa = 4.13EE165 pKa = 4.48DD166 pKa = 3.66IPRR169 pKa = 11.84HH170 pKa = 4.69AWQIALMKK178 pKa = 9.2EE179 pKa = 4.5TEE181 pKa = 4.22KK182 pKa = 11.07PPTKK186 pKa = 9.88KK187 pKa = 9.91QRR189 pKa = 11.84RR190 pKa = 11.84QIIWVVDD197 pKa = 3.67RR198 pKa = 11.84VGNTGKK204 pKa = 9.9SQLAKK209 pKa = 10.63YY210 pKa = 9.64MIMSDD215 pKa = 3.33PKK217 pKa = 10.49KK218 pKa = 9.99WFVCKK223 pKa = 10.55DD224 pKa = 3.18LGTSRR229 pKa = 11.84DD230 pKa = 3.44SSTIIQFAIAKK241 pKa = 8.34GWEE244 pKa = 4.01GHH246 pKa = 5.65GVIIDD251 pKa = 4.07LPRR254 pKa = 11.84SAEE257 pKa = 3.89NHH259 pKa = 5.88DD260 pKa = 3.55RR261 pKa = 11.84MYY263 pKa = 11.2SYY265 pKa = 10.66IEE267 pKa = 4.18EE268 pKa = 4.3IKK270 pKa = 11.0DD271 pKa = 3.85GFVTSQKK278 pKa = 10.42YY279 pKa = 8.68QGCTSTFNKK288 pKa = 9.18PHH290 pKa = 7.16LIVFANWEE298 pKa = 4.06PNYY301 pKa = 10.21TRR303 pKa = 11.84LSYY306 pKa = 10.83DD307 pKa = 2.55RR308 pKa = 11.84WDD310 pKa = 3.45MRR312 pKa = 11.84RR313 pKa = 11.84LEE315 pKa = 4.42VKK317 pKa = 10.4GKK319 pKa = 8.03TITMEE324 pKa = 4.08SFIPTILKK332 pKa = 10.09KK333 pKa = 10.46KK334 pKa = 8.79RR335 pKa = 11.84LEE337 pKa = 3.95YY338 pKa = 10.43HH339 pKa = 6.33AFDD342 pKa = 4.28YY343 pKa = 10.92QSQPDD348 pKa = 3.8EE349 pKa = 4.35DD350 pKa = 4.34HH351 pKa = 6.33LHH353 pKa = 6.89GYY355 pKa = 8.0ICKK358 pKa = 10.2EE359 pKa = 4.24LMDD362 pKa = 4.77DD363 pKa = 5.33LIGEE367 pKa = 4.31EE368 pKa = 4.64MLNEE372 pKa = 4.33HH373 pKa = 7.49DD374 pKa = 4.76PGCVDD379 pKa = 4.55EE380 pKa = 6.1IEE382 pKa = 4.52ATDD385 pKa = 4.44SEE387 pKa = 4.91VDD389 pKa = 3.76DD390 pKa = 5.02TPDD393 pKa = 3.45DD394 pKa = 4.05TPEE397 pKa = 3.9DD398 pKa = 3.97VYY400 pKa = 11.45DD401 pKa = 4.2IDD403 pKa = 5.95FDD405 pKa = 4.5DD406 pKa = 5.84LLVSDD411 pKa = 5.32DD412 pKa = 4.86LEE414 pKa = 4.19
MM1 pKa = 7.79ASTKK5 pKa = 10.25KK6 pKa = 10.46FRR8 pKa = 11.84FQNTSALLTYY18 pKa = 8.3KK19 pKa = 9.25THH21 pKa = 7.07LDD23 pKa = 3.04KK24 pKa = 10.96KK25 pKa = 10.89AYY27 pKa = 10.47LEE29 pKa = 4.12WFEE32 pKa = 4.61KK33 pKa = 9.95QAKK36 pKa = 5.67QTPVFIRR43 pKa = 11.84LAHH46 pKa = 5.47EE47 pKa = 4.52TGSHH51 pKa = 7.43DD52 pKa = 4.45EE53 pKa = 4.61GEE55 pKa = 4.05PDD57 pKa = 3.4YY58 pKa = 11.73DD59 pKa = 3.53HH60 pKa = 6.32THH62 pKa = 5.45VVFKK66 pKa = 10.48CKK68 pKa = 10.39SRR70 pKa = 11.84IDD72 pKa = 3.52TTSQRR77 pKa = 11.84FFDD80 pKa = 4.72FNAIHH85 pKa = 6.06PHH87 pKa = 5.17IKK89 pKa = 9.9VLKK92 pKa = 7.7TNKK95 pKa = 10.46SIDD98 pKa = 3.58DD99 pKa = 3.67AKK101 pKa = 11.08KK102 pKa = 10.78YY103 pKa = 9.13IAKK106 pKa = 9.67EE107 pKa = 4.0DD108 pKa = 3.86KK109 pKa = 10.91DD110 pKa = 4.45NEE112 pKa = 4.26DD113 pKa = 4.42LLVSDD118 pKa = 4.32TSNFIEE124 pKa = 5.0GVWSKK129 pKa = 9.28NTIQEE134 pKa = 4.06ALLDD138 pKa = 4.23FVKK141 pKa = 10.66KK142 pKa = 10.08PSDD145 pKa = 3.38VAGIIQLYY153 pKa = 8.88GLKK156 pKa = 10.41KK157 pKa = 10.56NPTQIHH163 pKa = 6.1EE164 pKa = 4.13EE165 pKa = 4.48DD166 pKa = 3.66IPRR169 pKa = 11.84HH170 pKa = 4.69AWQIALMKK178 pKa = 9.2EE179 pKa = 4.5TEE181 pKa = 4.22KK182 pKa = 11.07PPTKK186 pKa = 9.88KK187 pKa = 9.91QRR189 pKa = 11.84RR190 pKa = 11.84QIIWVVDD197 pKa = 3.67RR198 pKa = 11.84VGNTGKK204 pKa = 9.9SQLAKK209 pKa = 10.63YY210 pKa = 9.64MIMSDD215 pKa = 3.33PKK217 pKa = 10.49KK218 pKa = 9.99WFVCKK223 pKa = 10.55DD224 pKa = 3.18LGTSRR229 pKa = 11.84DD230 pKa = 3.44SSTIIQFAIAKK241 pKa = 8.34GWEE244 pKa = 4.01GHH246 pKa = 5.65GVIIDD251 pKa = 4.07LPRR254 pKa = 11.84SAEE257 pKa = 3.89NHH259 pKa = 5.88DD260 pKa = 3.55RR261 pKa = 11.84MYY263 pKa = 11.2SYY265 pKa = 10.66IEE267 pKa = 4.18EE268 pKa = 4.3IKK270 pKa = 11.0DD271 pKa = 3.85GFVTSQKK278 pKa = 10.42YY279 pKa = 8.68QGCTSTFNKK288 pKa = 9.18PHH290 pKa = 7.16LIVFANWEE298 pKa = 4.06PNYY301 pKa = 10.21TRR303 pKa = 11.84LSYY306 pKa = 10.83DD307 pKa = 2.55RR308 pKa = 11.84WDD310 pKa = 3.45MRR312 pKa = 11.84RR313 pKa = 11.84LEE315 pKa = 4.42VKK317 pKa = 10.4GKK319 pKa = 8.03TITMEE324 pKa = 4.08SFIPTILKK332 pKa = 10.09KK333 pKa = 10.46KK334 pKa = 8.79RR335 pKa = 11.84LEE337 pKa = 3.95YY338 pKa = 10.43HH339 pKa = 6.33AFDD342 pKa = 4.28YY343 pKa = 10.92QSQPDD348 pKa = 3.8EE349 pKa = 4.35DD350 pKa = 4.34HH351 pKa = 6.33LHH353 pKa = 6.89GYY355 pKa = 8.0ICKK358 pKa = 10.2EE359 pKa = 4.24LMDD362 pKa = 4.77DD363 pKa = 5.33LIGEE367 pKa = 4.31EE368 pKa = 4.64MLNEE372 pKa = 4.33HH373 pKa = 7.49DD374 pKa = 4.76PGCVDD379 pKa = 4.55EE380 pKa = 6.1IEE382 pKa = 4.52ATDD385 pKa = 4.44SEE387 pKa = 4.91VDD389 pKa = 3.76DD390 pKa = 5.02TPDD393 pKa = 3.45DD394 pKa = 4.05TPEE397 pKa = 3.9DD398 pKa = 3.97VYY400 pKa = 11.45DD401 pKa = 4.2IDD403 pKa = 5.95FDD405 pKa = 4.5DD406 pKa = 5.84LLVSDD411 pKa = 5.32DD412 pKa = 4.86LEE414 pKa = 4.19
Molecular weight: 48.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPB1|A0A346BPB1_9VIRU Putative capsid protein OS=Fire ant associated circular virus 1 OX=2293280 PE=3 SV=1
MM1 pKa = 7.64SGRR4 pKa = 11.84KK5 pKa = 7.77KK6 pKa = 8.83TASAPAKK13 pKa = 9.93KK14 pKa = 9.88YY15 pKa = 9.7YY16 pKa = 10.11RR17 pKa = 11.84YY18 pKa = 9.78AARR21 pKa = 11.84RR22 pKa = 11.84PAVVTGSGDD31 pKa = 3.42YY32 pKa = 10.29KK33 pKa = 10.01VARR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.64QKK40 pKa = 10.8SSMRR44 pKa = 11.84YY45 pKa = 7.76PGAGKK50 pKa = 8.8TIGSAIGSVFGPAGSLVGGALGNLAHH76 pKa = 6.71SALHH80 pKa = 6.92AITGFGDD87 pKa = 3.2YY88 pKa = 9.91HH89 pKa = 6.65VKK91 pKa = 10.75HH92 pKa = 6.07NVLTEE97 pKa = 4.32TNGPPQVINQGKK109 pKa = 8.97EE110 pKa = 3.4FIIRR114 pKa = 11.84HH115 pKa = 5.5RR116 pKa = 11.84EE117 pKa = 3.96YY118 pKa = 9.71ITDD121 pKa = 3.86IYY123 pKa = 11.12SSAGTASTPSPFLNQIFPINPGQATTFPWLASLAGKK159 pKa = 9.87FEE161 pKa = 4.23QYY163 pKa = 10.31RR164 pKa = 11.84VDD166 pKa = 3.51GMVFEE171 pKa = 4.98FKK173 pKa = 10.5SLYY176 pKa = 10.09SDD178 pKa = 4.52AVVTQNGSIGSIILATEE195 pKa = 4.15YY196 pKa = 10.86NAGSAPFNSKK206 pKa = 7.77QQMEE210 pKa = 4.77NYY212 pKa = 9.8QFAQSCKK219 pKa = 8.25PSLSVLHH226 pKa = 6.76PIEE229 pKa = 4.4CARR232 pKa = 11.84NQTVLSEE239 pKa = 4.39LYY241 pKa = 9.85VRR243 pKa = 11.84PSAVPTGEE251 pKa = 4.7DD252 pKa = 2.67IKK254 pKa = 10.24TYY256 pKa = 11.43DD257 pKa = 3.44MGDD260 pKa = 3.52FQIASQGIPLGSAGAPVNLGEE281 pKa = 4.8LWVTYY286 pKa = 9.89QISLLKK292 pKa = 10.36PKK294 pKa = 10.03IPSVGPASYY303 pKa = 11.08VDD305 pKa = 4.0SGWSHH310 pKa = 7.1FNTNNLVSGSFTAATPFGPLGSLFPLPQNNILGITFTTNSMSIPLTSTPMRR361 pKa = 11.84YY362 pKa = 8.99MIDD365 pKa = 3.46LWWTGDD371 pKa = 3.57TNVAGWNAPATGTITNGLLINAQIMTGSYY400 pKa = 10.19LIPNVGGSATTIGASAKK417 pKa = 10.54YY418 pKa = 9.95FIEE421 pKa = 4.78CPAATGSLNTCLIPIQNNGGFPSATIRR448 pKa = 11.84RR449 pKa = 11.84MFSS452 pKa = 2.7
MM1 pKa = 7.64SGRR4 pKa = 11.84KK5 pKa = 7.77KK6 pKa = 8.83TASAPAKK13 pKa = 9.93KK14 pKa = 9.88YY15 pKa = 9.7YY16 pKa = 10.11RR17 pKa = 11.84YY18 pKa = 9.78AARR21 pKa = 11.84RR22 pKa = 11.84PAVVTGSGDD31 pKa = 3.42YY32 pKa = 10.29KK33 pKa = 10.01VARR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.64QKK40 pKa = 10.8SSMRR44 pKa = 11.84YY45 pKa = 7.76PGAGKK50 pKa = 8.8TIGSAIGSVFGPAGSLVGGALGNLAHH76 pKa = 6.71SALHH80 pKa = 6.92AITGFGDD87 pKa = 3.2YY88 pKa = 9.91HH89 pKa = 6.65VKK91 pKa = 10.75HH92 pKa = 6.07NVLTEE97 pKa = 4.32TNGPPQVINQGKK109 pKa = 8.97EE110 pKa = 3.4FIIRR114 pKa = 11.84HH115 pKa = 5.5RR116 pKa = 11.84EE117 pKa = 3.96YY118 pKa = 9.71ITDD121 pKa = 3.86IYY123 pKa = 11.12SSAGTASTPSPFLNQIFPINPGQATTFPWLASLAGKK159 pKa = 9.87FEE161 pKa = 4.23QYY163 pKa = 10.31RR164 pKa = 11.84VDD166 pKa = 3.51GMVFEE171 pKa = 4.98FKK173 pKa = 10.5SLYY176 pKa = 10.09SDD178 pKa = 4.52AVVTQNGSIGSIILATEE195 pKa = 4.15YY196 pKa = 10.86NAGSAPFNSKK206 pKa = 7.77QQMEE210 pKa = 4.77NYY212 pKa = 9.8QFAQSCKK219 pKa = 8.25PSLSVLHH226 pKa = 6.76PIEE229 pKa = 4.4CARR232 pKa = 11.84NQTVLSEE239 pKa = 4.39LYY241 pKa = 9.85VRR243 pKa = 11.84PSAVPTGEE251 pKa = 4.7DD252 pKa = 2.67IKK254 pKa = 10.24TYY256 pKa = 11.43DD257 pKa = 3.44MGDD260 pKa = 3.52FQIASQGIPLGSAGAPVNLGEE281 pKa = 4.8LWVTYY286 pKa = 9.89QISLLKK292 pKa = 10.36PKK294 pKa = 10.03IPSVGPASYY303 pKa = 11.08VDD305 pKa = 4.0SGWSHH310 pKa = 7.1FNTNNLVSGSFTAATPFGPLGSLFPLPQNNILGITFTTNSMSIPLTSTPMRR361 pKa = 11.84YY362 pKa = 8.99MIDD365 pKa = 3.46LWWTGDD371 pKa = 3.57TNVAGWNAPATGTITNGLLINAQIMTGSYY400 pKa = 10.19LIPNVGGSATTIGASAKK417 pKa = 10.54YY418 pKa = 9.95FIEE421 pKa = 4.78CPAATGSLNTCLIPIQNNGGFPSATIRR448 pKa = 11.84RR449 pKa = 11.84MFSS452 pKa = 2.7
Molecular weight: 48.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
866 |
414 |
452 |
433.0 |
48.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.044 ± 1.606 | 1.039 ± 0.11 |
6.236 ± 2.716 | 4.965 ± 1.651 |
4.388 ± 0.026 | 7.275 ± 2.073 |
2.656 ± 0.791 | 7.275 ± 0.139 |
6.697 ± 1.94 | 6.813 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.194 ± 0.013 | 4.503 ± 0.892 |
5.658 ± 1.015 | 3.926 ± 0.04 |
3.58 ± 0.187 | 7.852 ± 1.345 |
7.39 ± 0.41 | 4.965 ± 0.088 |
1.617 ± 0.207 | 3.926 ± 0.198 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
