
Smithella sp. ME-1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Syntrophobacterales; Syntrophaceae; Smithella; unclassified Smithella
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
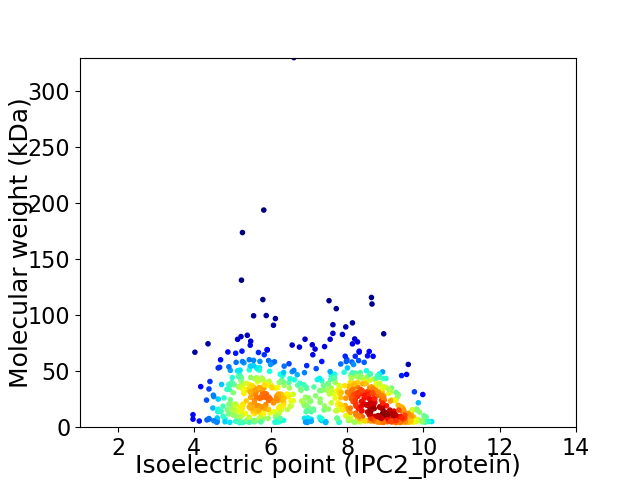
Virtual 2D-PAGE plot for 705 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
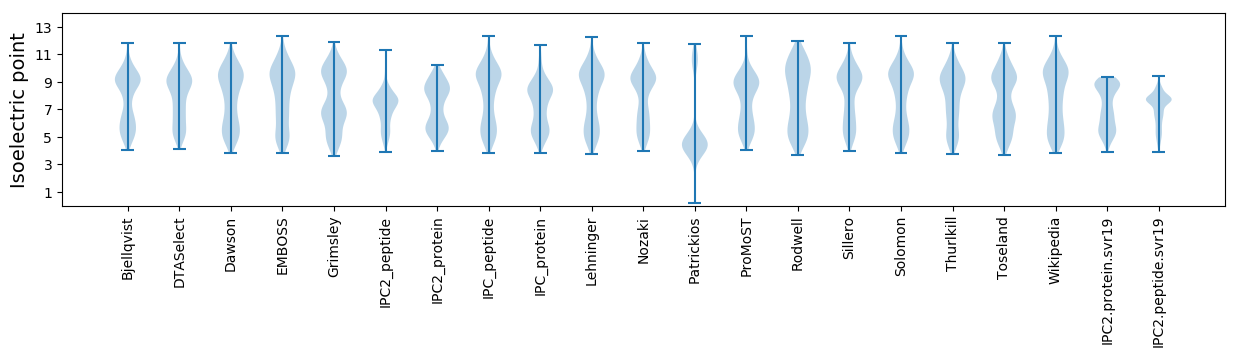
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V4F9J5|V4F9J5_9DELT Uncharacterized protein OS=Smithella sp. ME-1 OX=1380771 GN=SMITH_272 PE=4 SV=1
MM1 pKa = 7.43ILSACSNSVLTPAPPQQPSSTSEE24 pKa = 3.76PTKK27 pKa = 10.91EE28 pKa = 4.54EE29 pKa = 4.01IIEE32 pKa = 4.18PNALPTLMATEE43 pKa = 4.42TTVPATPTLEE53 pKa = 3.95PTIVPTEE60 pKa = 3.91VGDD63 pKa = 3.72PNFRR67 pKa = 11.84ILEE70 pKa = 4.27KK71 pKa = 10.76DD72 pKa = 2.91QSEE75 pKa = 4.55QIFIPAGEE83 pKa = 4.82FIMGTQDD90 pKa = 2.51TDD92 pKa = 3.5AQHH95 pKa = 6.33TLDD98 pKa = 4.45NGVAYY103 pKa = 9.63PEE105 pKa = 4.48VPMHH109 pKa = 5.96TVYY112 pKa = 11.2LDD114 pKa = 3.84GYY116 pKa = 9.16WIDD119 pKa = 3.84KK120 pKa = 11.14YY121 pKa = 11.09EE122 pKa = 4.18VTNGRR127 pKa = 11.84YY128 pKa = 8.95ALCVADD134 pKa = 5.09GACTAPWVNFSNTRR148 pKa = 11.84QEE150 pKa = 4.51YY151 pKa = 9.17YY152 pKa = 11.15GNPEE156 pKa = 3.71FDD158 pKa = 3.78NYY160 pKa = 9.38PVIMVDD166 pKa = 2.51WWQSNDD172 pKa = 2.87FCEE175 pKa = 4.39WAGGRR180 pKa = 11.84LPTEE184 pKa = 4.47AEE186 pKa = 3.75WEE188 pKa = 4.16KK189 pKa = 10.84AARR192 pKa = 11.84GTDD195 pKa = 3.1GRR197 pKa = 11.84KK198 pKa = 9.55YY199 pKa = 10.29PWGNDD204 pKa = 2.98PLTEE208 pKa = 4.65DD209 pKa = 3.89KK210 pKa = 11.64ANFCDD215 pKa = 4.25VNCTRR220 pKa = 11.84EE221 pKa = 4.03HH222 pKa = 6.84ANPAFDD228 pKa = 6.01DD229 pKa = 4.56GFADD233 pKa = 3.99TAPVGSYY240 pKa = 10.89PNGASPYY247 pKa = 10.44GVMDD251 pKa = 3.24MAGNVWEE258 pKa = 4.39WTGSIPKK265 pKa = 9.43PYY267 pKa = 9.97PYY269 pKa = 10.74DD270 pKa = 4.5PDD272 pKa = 4.07DD273 pKa = 4.05GRR275 pKa = 11.84EE276 pKa = 3.96EE277 pKa = 4.29NDD279 pKa = 3.36GYY281 pKa = 11.24LYY283 pKa = 10.62VWRR286 pKa = 11.84GGPWSNGTWWIRR298 pKa = 11.84STLRR302 pKa = 11.84YY303 pKa = 9.82KK304 pKa = 10.48SVPKK308 pKa = 10.13YY309 pKa = 9.47WYY311 pKa = 7.99EE312 pKa = 3.85NLGFRR317 pKa = 11.84CASTDD322 pKa = 2.96
MM1 pKa = 7.43ILSACSNSVLTPAPPQQPSSTSEE24 pKa = 3.76PTKK27 pKa = 10.91EE28 pKa = 4.54EE29 pKa = 4.01IIEE32 pKa = 4.18PNALPTLMATEE43 pKa = 4.42TTVPATPTLEE53 pKa = 3.95PTIVPTEE60 pKa = 3.91VGDD63 pKa = 3.72PNFRR67 pKa = 11.84ILEE70 pKa = 4.27KK71 pKa = 10.76DD72 pKa = 2.91QSEE75 pKa = 4.55QIFIPAGEE83 pKa = 4.82FIMGTQDD90 pKa = 2.51TDD92 pKa = 3.5AQHH95 pKa = 6.33TLDD98 pKa = 4.45NGVAYY103 pKa = 9.63PEE105 pKa = 4.48VPMHH109 pKa = 5.96TVYY112 pKa = 11.2LDD114 pKa = 3.84GYY116 pKa = 9.16WIDD119 pKa = 3.84KK120 pKa = 11.14YY121 pKa = 11.09EE122 pKa = 4.18VTNGRR127 pKa = 11.84YY128 pKa = 8.95ALCVADD134 pKa = 5.09GACTAPWVNFSNTRR148 pKa = 11.84QEE150 pKa = 4.51YY151 pKa = 9.17YY152 pKa = 11.15GNPEE156 pKa = 3.71FDD158 pKa = 3.78NYY160 pKa = 9.38PVIMVDD166 pKa = 2.51WWQSNDD172 pKa = 2.87FCEE175 pKa = 4.39WAGGRR180 pKa = 11.84LPTEE184 pKa = 4.47AEE186 pKa = 3.75WEE188 pKa = 4.16KK189 pKa = 10.84AARR192 pKa = 11.84GTDD195 pKa = 3.1GRR197 pKa = 11.84KK198 pKa = 9.55YY199 pKa = 10.29PWGNDD204 pKa = 2.98PLTEE208 pKa = 4.65DD209 pKa = 3.89KK210 pKa = 11.64ANFCDD215 pKa = 4.25VNCTRR220 pKa = 11.84EE221 pKa = 4.03HH222 pKa = 6.84ANPAFDD228 pKa = 6.01DD229 pKa = 4.56GFADD233 pKa = 3.99TAPVGSYY240 pKa = 10.89PNGASPYY247 pKa = 10.44GVMDD251 pKa = 3.24MAGNVWEE258 pKa = 4.39WTGSIPKK265 pKa = 9.43PYY267 pKa = 9.97PYY269 pKa = 10.74DD270 pKa = 4.5PDD272 pKa = 4.07DD273 pKa = 4.05GRR275 pKa = 11.84EE276 pKa = 3.96EE277 pKa = 4.29NDD279 pKa = 3.36GYY281 pKa = 11.24LYY283 pKa = 10.62VWRR286 pKa = 11.84GGPWSNGTWWIRR298 pKa = 11.84STLRR302 pKa = 11.84YY303 pKa = 9.82KK304 pKa = 10.48SVPKK308 pKa = 10.13YY309 pKa = 9.47WYY311 pKa = 7.99EE312 pKa = 3.85NLGFRR317 pKa = 11.84CASTDD322 pKa = 2.96
Molecular weight: 36.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V4GT14|V4GT14_9DELT Oxygen-insensitive NADPH nitroreductase OS=Smithella sp. ME-1 OX=1380771 GN=SMITH_144 PE=4 SV=1
MM1 pKa = 7.38MNHH4 pKa = 7.03ILNATLLVALIVAFAAGLALGAFYY28 pKa = 10.03FIALWHH34 pKa = 5.5TLQRR38 pKa = 11.84LPAAHH43 pKa = 6.33SPARR47 pKa = 11.84LMLVSFVLRR56 pKa = 11.84MAVVMVGFYY65 pKa = 10.74SIMGGEE71 pKa = 3.59HH72 pKa = 6.13WEE74 pKa = 4.03RR75 pKa = 11.84LVAAMLGFIIIRR87 pKa = 11.84KK88 pKa = 8.98ILTHH92 pKa = 6.76LLGPQKK98 pKa = 9.63MVEE101 pKa = 4.09EE102 pKa = 4.48VRR104 pKa = 11.84CINLGRR110 pKa = 4.05
MM1 pKa = 7.38MNHH4 pKa = 7.03ILNATLLVALIVAFAAGLALGAFYY28 pKa = 10.03FIALWHH34 pKa = 5.5TLQRR38 pKa = 11.84LPAAHH43 pKa = 6.33SPARR47 pKa = 11.84LMLVSFVLRR56 pKa = 11.84MAVVMVGFYY65 pKa = 10.74SIMGGEE71 pKa = 3.59HH72 pKa = 6.13WEE74 pKa = 4.03RR75 pKa = 11.84LVAAMLGFIIIRR87 pKa = 11.84KK88 pKa = 8.98ILTHH92 pKa = 6.76LLGPQKK98 pKa = 9.63MVEE101 pKa = 4.09EE102 pKa = 4.48VRR104 pKa = 11.84CINLGRR110 pKa = 4.05
Molecular weight: 12.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
183521 |
37 |
2904 |
260.3 |
29.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.59 ± 0.099 | 1.248 ± 0.042 |
5.255 ± 0.065 | 6.379 ± 0.09 |
4.599 ± 0.075 | 6.729 ± 0.085 |
1.825 ± 0.042 | 8.301 ± 0.099 |
7.392 ± 0.128 | 9.439 ± 0.104 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.714 ± 0.043 | 4.592 ± 0.073 |
3.997 ± 0.062 | 3.197 ± 0.052 |
4.77 ± 0.087 | 5.982 ± 0.073 |
4.973 ± 0.062 | 6.545 ± 0.079 |
1.101 ± 0.039 | 3.37 ± 0.059 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
