
Lake Sarah-associated circular virus-31
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.93
Get precalculated fractions of proteins
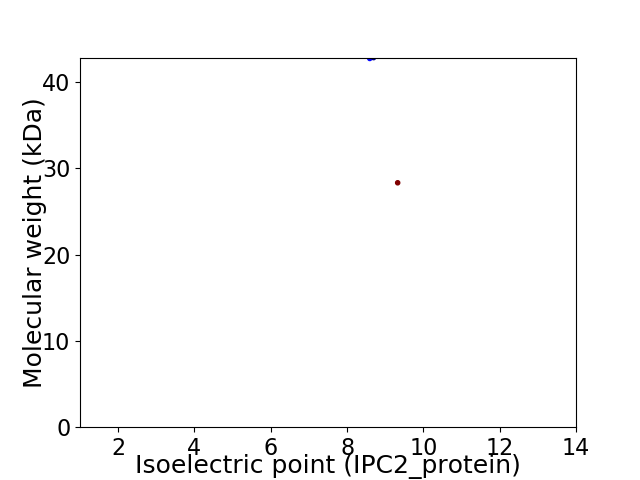
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQN6|A0A140AQN6_9VIRU Coat protein OS=Lake Sarah-associated circular virus-31 OX=1685759 PE=4 SV=1
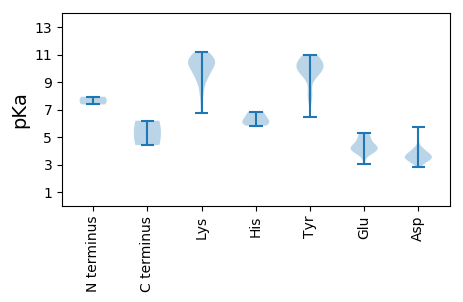
MM1 pKa = 7.37SRR3 pKa = 11.84SQAYY7 pKa = 9.93CFTVNNYY14 pKa = 9.58EE15 pKa = 5.02GLLDD19 pKa = 4.03WEE21 pKa = 4.77DD22 pKa = 3.71LQLAGATYY30 pKa = 10.6LIYY33 pKa = 10.37QEE35 pKa = 5.15EE36 pKa = 4.39IGEE39 pKa = 4.13QGTIHH44 pKa = 6.12LQGYY48 pKa = 9.93IYY50 pKa = 10.65FSTKK54 pKa = 10.18KK55 pKa = 10.65SMKK58 pKa = 9.47QLCALIDD65 pKa = 4.04GSSVRR70 pKa = 11.84VANGSPQQNTTYY82 pKa = 10.33CSKK85 pKa = 11.14EE86 pKa = 3.86EE87 pKa = 4.0GRR89 pKa = 11.84LGGPYY94 pKa = 10.12VYY96 pKa = 9.89GTMPQQVRR104 pKa = 11.84TTSIASAASTARR116 pKa = 11.84GDD118 pKa = 3.6KK119 pKa = 10.39NKK121 pKa = 10.34KK122 pKa = 6.76EE123 pKa = 4.24RR124 pKa = 11.84KK125 pKa = 9.2GEE127 pKa = 3.94EE128 pKa = 3.66FSRR131 pKa = 11.84ANSKK135 pKa = 10.74KK136 pKa = 10.44NYY138 pKa = 7.98SAKK141 pKa = 10.34NFYY144 pKa = 9.87PPRR147 pKa = 11.84EE148 pKa = 4.13KK149 pKa = 10.81EE150 pKa = 4.32PTCSPSKK157 pKa = 9.09PIKK160 pKa = 10.13MPACPKK166 pKa = 9.64PNSGTITMPICALSQEE182 pKa = 4.09RR183 pKa = 11.84KK184 pKa = 9.47IYY186 pKa = 10.69SEE188 pKa = 4.71LKK190 pKa = 9.56IKK192 pKa = 9.87PRR194 pKa = 11.84NTKK197 pKa = 8.72PTVLLLVGPSGTGKK211 pKa = 10.35SRR213 pKa = 11.84TAYY216 pKa = 10.31QLAQFLGLTIYY227 pKa = 9.66VVPPTKK233 pKa = 10.41GGSGLRR239 pKa = 11.84FDD241 pKa = 5.75GYY243 pKa = 10.83NQQDD247 pKa = 3.4VCIIDD252 pKa = 4.03EE253 pKa = 4.28VDD255 pKa = 3.44GSSFTPTFFNLLCDD269 pKa = 3.78WYY271 pKa = 9.3EE272 pKa = 3.93MKK274 pKa = 10.99VPVHH278 pKa = 5.83GTANLQFTSPYY289 pKa = 10.2IILCSNYY296 pKa = 9.72LPKK299 pKa = 10.61YY300 pKa = 6.42WWKK303 pKa = 10.59SRR305 pKa = 11.84SRR307 pKa = 11.84DD308 pKa = 3.44QVKK311 pKa = 8.1QTTRR315 pKa = 11.84RR316 pKa = 11.84IDD318 pKa = 2.91ITIPFFRR325 pKa = 11.84RR326 pKa = 11.84TPQVQPAPSINLPEE340 pKa = 5.21DD341 pKa = 3.5LDD343 pKa = 3.75EE344 pKa = 5.27FGFVTVHH351 pKa = 6.85PIKK354 pKa = 10.08MPHH357 pKa = 6.53YY358 pKa = 8.1YY359 pKa = 9.91VQPLEE364 pKa = 4.2RR365 pKa = 11.84FPPKK369 pKa = 9.65ITDD372 pKa = 3.25KK373 pKa = 10.86MIFPLL378 pKa = 4.41
MM1 pKa = 7.37SRR3 pKa = 11.84SQAYY7 pKa = 9.93CFTVNNYY14 pKa = 9.58EE15 pKa = 5.02GLLDD19 pKa = 4.03WEE21 pKa = 4.77DD22 pKa = 3.71LQLAGATYY30 pKa = 10.6LIYY33 pKa = 10.37QEE35 pKa = 5.15EE36 pKa = 4.39IGEE39 pKa = 4.13QGTIHH44 pKa = 6.12LQGYY48 pKa = 9.93IYY50 pKa = 10.65FSTKK54 pKa = 10.18KK55 pKa = 10.65SMKK58 pKa = 9.47QLCALIDD65 pKa = 4.04GSSVRR70 pKa = 11.84VANGSPQQNTTYY82 pKa = 10.33CSKK85 pKa = 11.14EE86 pKa = 3.86EE87 pKa = 4.0GRR89 pKa = 11.84LGGPYY94 pKa = 10.12VYY96 pKa = 9.89GTMPQQVRR104 pKa = 11.84TTSIASAASTARR116 pKa = 11.84GDD118 pKa = 3.6KK119 pKa = 10.39NKK121 pKa = 10.34KK122 pKa = 6.76EE123 pKa = 4.24RR124 pKa = 11.84KK125 pKa = 9.2GEE127 pKa = 3.94EE128 pKa = 3.66FSRR131 pKa = 11.84ANSKK135 pKa = 10.74KK136 pKa = 10.44NYY138 pKa = 7.98SAKK141 pKa = 10.34NFYY144 pKa = 9.87PPRR147 pKa = 11.84EE148 pKa = 4.13KK149 pKa = 10.81EE150 pKa = 4.32PTCSPSKK157 pKa = 9.09PIKK160 pKa = 10.13MPACPKK166 pKa = 9.64PNSGTITMPICALSQEE182 pKa = 4.09RR183 pKa = 11.84KK184 pKa = 9.47IYY186 pKa = 10.69SEE188 pKa = 4.71LKK190 pKa = 9.56IKK192 pKa = 9.87PRR194 pKa = 11.84NTKK197 pKa = 8.72PTVLLLVGPSGTGKK211 pKa = 10.35SRR213 pKa = 11.84TAYY216 pKa = 10.31QLAQFLGLTIYY227 pKa = 9.66VVPPTKK233 pKa = 10.41GGSGLRR239 pKa = 11.84FDD241 pKa = 5.75GYY243 pKa = 10.83NQQDD247 pKa = 3.4VCIIDD252 pKa = 4.03EE253 pKa = 4.28VDD255 pKa = 3.44GSSFTPTFFNLLCDD269 pKa = 3.78WYY271 pKa = 9.3EE272 pKa = 3.93MKK274 pKa = 10.99VPVHH278 pKa = 5.83GTANLQFTSPYY289 pKa = 10.2IILCSNYY296 pKa = 9.72LPKK299 pKa = 10.61YY300 pKa = 6.42WWKK303 pKa = 10.59SRR305 pKa = 11.84SRR307 pKa = 11.84DD308 pKa = 3.44QVKK311 pKa = 8.1QTTRR315 pKa = 11.84RR316 pKa = 11.84IDD318 pKa = 2.91ITIPFFRR325 pKa = 11.84RR326 pKa = 11.84TPQVQPAPSINLPEE340 pKa = 5.21DD341 pKa = 3.5LDD343 pKa = 3.75EE344 pKa = 5.27FGFVTVHH351 pKa = 6.85PIKK354 pKa = 10.08MPHH357 pKa = 6.53YY358 pKa = 8.1YY359 pKa = 9.91VQPLEE364 pKa = 4.2RR365 pKa = 11.84FPPKK369 pKa = 9.65ITDD372 pKa = 3.25KK373 pKa = 10.86MIFPLL378 pKa = 4.41
Molecular weight: 42.74 kDa
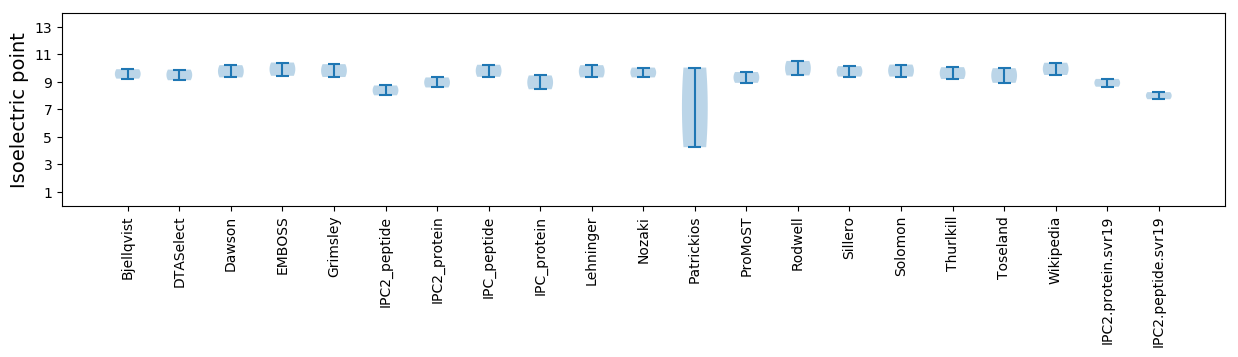
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQN6|A0A140AQN6_9VIRU Coat protein OS=Lake Sarah-associated circular virus-31 OX=1685759 PE=4 SV=1
MM1 pKa = 7.93LSRR4 pKa = 11.84KK5 pKa = 8.61DD6 pKa = 3.37LKK8 pKa = 11.17KK9 pKa = 10.37IVTKK13 pKa = 10.41RR14 pKa = 11.84ASSKK18 pKa = 10.88ASAASKK24 pKa = 10.23AARR27 pKa = 11.84SSNMSARR34 pKa = 11.84RR35 pKa = 11.84GPAPTRR41 pKa = 11.84QAQYY45 pKa = 10.12ASAVSRR51 pKa = 11.84SFGSGRR57 pKa = 11.84EE58 pKa = 3.93NKK60 pKa = 9.42TFDD63 pKa = 3.67AVVLGTNILRR73 pKa = 11.84ASTDD77 pKa = 3.32DD78 pKa = 3.46AVAISASGYY87 pKa = 7.99ITTNSSAMVLNQVPQSTTSTTRR109 pKa = 11.84IGRR112 pKa = 11.84KK113 pKa = 7.7MVMKK117 pKa = 10.83GILLQGFIAAGQTGSLGSLARR138 pKa = 11.84MCLVYY143 pKa = 10.29IPRR146 pKa = 11.84LDD148 pKa = 3.51RR149 pKa = 11.84TTTTMPPQNVIWSAQNPCALRR170 pKa = 11.84VINNSDD176 pKa = 3.33RR177 pKa = 11.84FRR179 pKa = 11.84IIRR182 pKa = 11.84QWTHH186 pKa = 6.1KK187 pKa = 9.97LTGDD191 pKa = 3.97FDD193 pKa = 4.31AATTGNEE200 pKa = 3.01IVAFSEE206 pKa = 4.39YY207 pKa = 11.0VKK209 pKa = 11.07LDD211 pKa = 3.72LQTSWIQSNSDD222 pKa = 2.82GSFNDD227 pKa = 3.46MEE229 pKa = 4.58EE230 pKa = 4.42GALCLYY236 pKa = 10.64AQGLYY241 pKa = 9.83TGASGSAPTINFASRR256 pKa = 11.84IYY258 pKa = 10.87FEE260 pKa = 4.96DD261 pKa = 3.55HH262 pKa = 6.19
MM1 pKa = 7.93LSRR4 pKa = 11.84KK5 pKa = 8.61DD6 pKa = 3.37LKK8 pKa = 11.17KK9 pKa = 10.37IVTKK13 pKa = 10.41RR14 pKa = 11.84ASSKK18 pKa = 10.88ASAASKK24 pKa = 10.23AARR27 pKa = 11.84SSNMSARR34 pKa = 11.84RR35 pKa = 11.84GPAPTRR41 pKa = 11.84QAQYY45 pKa = 10.12ASAVSRR51 pKa = 11.84SFGSGRR57 pKa = 11.84EE58 pKa = 3.93NKK60 pKa = 9.42TFDD63 pKa = 3.67AVVLGTNILRR73 pKa = 11.84ASTDD77 pKa = 3.32DD78 pKa = 3.46AVAISASGYY87 pKa = 7.99ITTNSSAMVLNQVPQSTTSTTRR109 pKa = 11.84IGRR112 pKa = 11.84KK113 pKa = 7.7MVMKK117 pKa = 10.83GILLQGFIAAGQTGSLGSLARR138 pKa = 11.84MCLVYY143 pKa = 10.29IPRR146 pKa = 11.84LDD148 pKa = 3.51RR149 pKa = 11.84TTTTMPPQNVIWSAQNPCALRR170 pKa = 11.84VINNSDD176 pKa = 3.33RR177 pKa = 11.84FRR179 pKa = 11.84IIRR182 pKa = 11.84QWTHH186 pKa = 6.1KK187 pKa = 9.97LTGDD191 pKa = 3.97FDD193 pKa = 4.31AATTGNEE200 pKa = 3.01IVAFSEE206 pKa = 4.39YY207 pKa = 11.0VKK209 pKa = 11.07LDD211 pKa = 3.72LQTSWIQSNSDD222 pKa = 2.82GSFNDD227 pKa = 3.46MEE229 pKa = 4.58EE230 pKa = 4.42GALCLYY236 pKa = 10.64AQGLYY241 pKa = 9.83TGASGSAPTINFASRR256 pKa = 11.84IYY258 pKa = 10.87FEE260 pKa = 4.96DD261 pKa = 3.55HH262 pKa = 6.19
Molecular weight: 28.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
640 |
262 |
378 |
320.0 |
35.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.344 ± 2.565 | 1.875 ± 0.456 |
4.063 ± 0.323 | 3.906 ± 1.009 |
3.906 ± 0.294 | 6.719 ± 0.095 |
0.938 ± 0.109 | 6.094 ± 0.008 |
6.094 ± 1.184 | 7.031 ± 0.101 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 0.346 | 4.375 ± 0.366 |
6.25 ± 1.996 | 5.156 ± 0.36 |
5.938 ± 0.821 | 9.375 ± 1.534 |
8.125 ± 0.408 | 4.844 ± 0.074 |
1.094 ± 0.032 | 4.375 ± 1.064 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
