
Natronomonas pharaonis (strain ATCC 35678 / DSM 2160 / CIP 103997 / JCM 8858 / NBRC 14720 / NCIMB 2260 / Gabara) (Halobacterium pharaonis)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Natronomonas; Natronomonas pharaonis
Average proteome isoelectric point is 4.81
Get precalculated fractions of proteins
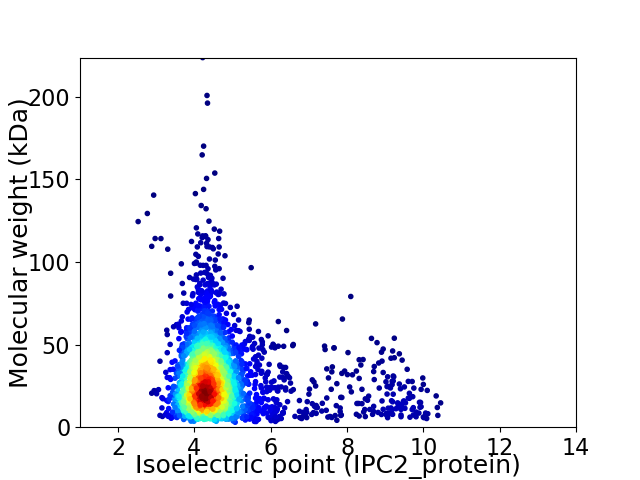
Virtual 2D-PAGE plot for 2764 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U7EY20|A0A1U7EY20_NATPD Uncharacterized protein OS=Natronomonas pharaonis (strain ATCC 35678 / DSM 2160 / CIP 103997 / JCM 8858 / NBRC 14720 / NCIMB 2260 / Gabara) OX=348780 GN=NP_3996A PE=4 SV=1
MM1 pKa = 7.51SPDD4 pKa = 3.22QSGSLSRR11 pKa = 11.84RR12 pKa = 11.84TVLVSAVAGGIGAVAGCLDD31 pKa = 4.15GDD33 pKa = 4.05RR34 pKa = 11.84DD35 pKa = 4.14GPDD38 pKa = 3.68PVSLDD43 pKa = 3.99DD44 pKa = 5.08GQACDD49 pKa = 3.5TCDD52 pKa = 3.59MLIEE56 pKa = 4.04MHH58 pKa = 7.07PGPVGQAFYY67 pKa = 11.3GDD69 pKa = 3.94DD70 pKa = 4.11APRR73 pKa = 11.84SLPDD77 pKa = 4.49DD78 pKa = 4.77RR79 pKa = 11.84DD80 pKa = 3.94DD81 pKa = 3.93GAAWFCSSTCAYY93 pKa = 10.44QFILEE98 pKa = 4.15EE99 pKa = 4.33EE100 pKa = 4.43EE101 pKa = 4.64RR102 pKa = 11.84GHH104 pKa = 6.36EE105 pKa = 4.0PAISYY110 pKa = 7.67GTDD113 pKa = 3.13YY114 pKa = 10.99STVDD118 pKa = 3.06YY119 pKa = 10.21TLRR122 pKa = 11.84DD123 pKa = 3.92DD124 pKa = 4.54NGVTVISAHH133 pKa = 6.47LEE135 pKa = 3.67ADD137 pKa = 3.62AFADD141 pKa = 3.68LHH143 pKa = 6.9DD144 pKa = 4.24LTFVADD150 pKa = 3.88SDD152 pKa = 4.76AEE154 pKa = 4.27GAMGGSLIGFSDD166 pKa = 4.03PDD168 pKa = 3.61DD169 pKa = 4.97AEE171 pKa = 4.66AFQSEE176 pKa = 4.61HH177 pKa = 6.92GGEE180 pKa = 4.08LLAHH184 pKa = 6.81DD185 pKa = 4.07EE186 pKa = 4.36VTRR189 pKa = 11.84EE190 pKa = 3.96VLTGLGMGMM199 pKa = 4.51
MM1 pKa = 7.51SPDD4 pKa = 3.22QSGSLSRR11 pKa = 11.84RR12 pKa = 11.84TVLVSAVAGGIGAVAGCLDD31 pKa = 4.15GDD33 pKa = 4.05RR34 pKa = 11.84DD35 pKa = 4.14GPDD38 pKa = 3.68PVSLDD43 pKa = 3.99DD44 pKa = 5.08GQACDD49 pKa = 3.5TCDD52 pKa = 3.59MLIEE56 pKa = 4.04MHH58 pKa = 7.07PGPVGQAFYY67 pKa = 11.3GDD69 pKa = 3.94DD70 pKa = 4.11APRR73 pKa = 11.84SLPDD77 pKa = 4.49DD78 pKa = 4.77RR79 pKa = 11.84DD80 pKa = 3.94DD81 pKa = 3.93GAAWFCSSTCAYY93 pKa = 10.44QFILEE98 pKa = 4.15EE99 pKa = 4.33EE100 pKa = 4.43EE101 pKa = 4.64RR102 pKa = 11.84GHH104 pKa = 6.36EE105 pKa = 4.0PAISYY110 pKa = 7.67GTDD113 pKa = 3.13YY114 pKa = 10.99STVDD118 pKa = 3.06YY119 pKa = 10.21TLRR122 pKa = 11.84DD123 pKa = 3.92DD124 pKa = 4.54NGVTVISAHH133 pKa = 6.47LEE135 pKa = 3.67ADD137 pKa = 3.62AFADD141 pKa = 3.68LHH143 pKa = 6.9DD144 pKa = 4.24LTFVADD150 pKa = 3.88SDD152 pKa = 4.76AEE154 pKa = 4.27GAMGGSLIGFSDD166 pKa = 4.03PDD168 pKa = 3.61DD169 pKa = 4.97AEE171 pKa = 4.66AFQSEE176 pKa = 4.61HH177 pKa = 6.92GGEE180 pKa = 4.08LLAHH184 pKa = 6.81DD185 pKa = 4.07EE186 pKa = 4.36VTRR189 pKa = 11.84EE190 pKa = 3.96VLTGLGMGMM199 pKa = 4.51
Molecular weight: 20.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U7EY34|A0A1U7EY34_NATPD DUF2339 family protein OS=Natronomonas pharaonis (strain ATCC 35678 / DSM 2160 / CIP 103997 / JCM 8858 / NBRC 14720 / NCIMB 2260 / Gabara) OX=348780 GN=NP_4038A PE=4 SV=1
MM1 pKa = 7.56NDD3 pKa = 3.67DD4 pKa = 4.09DD5 pKa = 5.68RR6 pKa = 11.84SPGMLSGGLVVPGLLGGLLVVYY28 pKa = 9.34FLLPFVAFLSRR39 pKa = 11.84AGSVDD44 pKa = 3.57VVAHH48 pKa = 6.41LSSPAARR55 pKa = 11.84RR56 pKa = 11.84AVRR59 pKa = 11.84NSLLTAPVATAIATGLGVPLAYY81 pKa = 10.76LLARR85 pKa = 11.84GSFRR89 pKa = 11.84GKK91 pKa = 10.22RR92 pKa = 11.84LLEE95 pKa = 4.16AAVVLPLVAPPVVGGVMLLTAVGQFTPLGRR125 pKa = 11.84VAASLGVPLTDD136 pKa = 3.17SLLGVVLAQLFVAAPFVILTARR158 pKa = 11.84AGFAAIDD165 pKa = 3.75EE166 pKa = 4.49RR167 pKa = 11.84LEE169 pKa = 3.87QASRR173 pKa = 11.84SLGYY177 pKa = 10.88GPIATFWRR185 pKa = 11.84VSLPLASGAIVVGVTLTFVRR205 pKa = 11.84AVGEE209 pKa = 4.57FGATMMVANNPRR221 pKa = 11.84TMPTQIWVDD230 pKa = 4.89FVAGNIDD237 pKa = 4.14GIVPLTLALLAVTLAVVFIVQRR259 pKa = 11.84LGRR262 pKa = 11.84TPAVVDD268 pKa = 3.54AA269 pKa = 5.24
MM1 pKa = 7.56NDD3 pKa = 3.67DD4 pKa = 4.09DD5 pKa = 5.68RR6 pKa = 11.84SPGMLSGGLVVPGLLGGLLVVYY28 pKa = 9.34FLLPFVAFLSRR39 pKa = 11.84AGSVDD44 pKa = 3.57VVAHH48 pKa = 6.41LSSPAARR55 pKa = 11.84RR56 pKa = 11.84AVRR59 pKa = 11.84NSLLTAPVATAIATGLGVPLAYY81 pKa = 10.76LLARR85 pKa = 11.84GSFRR89 pKa = 11.84GKK91 pKa = 10.22RR92 pKa = 11.84LLEE95 pKa = 4.16AAVVLPLVAPPVVGGVMLLTAVGQFTPLGRR125 pKa = 11.84VAASLGVPLTDD136 pKa = 3.17SLLGVVLAQLFVAAPFVILTARR158 pKa = 11.84AGFAAIDD165 pKa = 3.75EE166 pKa = 4.49RR167 pKa = 11.84LEE169 pKa = 3.87QASRR173 pKa = 11.84SLGYY177 pKa = 10.88GPIATFWRR185 pKa = 11.84VSLPLASGAIVVGVTLTFVRR205 pKa = 11.84AVGEE209 pKa = 4.57FGATMMVANNPRR221 pKa = 11.84TMPTQIWVDD230 pKa = 4.89FVAGNIDD237 pKa = 4.14GIVPLTLALLAVTLAVVFIVQRR259 pKa = 11.84LGRR262 pKa = 11.84TPAVVDD268 pKa = 3.54AA269 pKa = 5.24
Molecular weight: 27.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
810134 |
29 |
1999 |
293.1 |
31.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.416 ± 0.077 | 0.773 ± 0.016 |
8.745 ± 0.054 | 8.847 ± 0.06 |
3.253 ± 0.032 | 8.405 ± 0.041 |
1.97 ± 0.025 | 4.2 ± 0.033 |
1.891 ± 0.028 | 8.779 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.734 ± 0.021 | 2.211 ± 0.024 |
4.604 ± 0.035 | 2.584 ± 0.026 |
6.37 ± 0.05 | 5.255 ± 0.035 |
6.372 ± 0.034 | 8.893 ± 0.049 |
1.055 ± 0.017 | 2.643 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
