
Alethinophid 1 reptarenavirus (isolate AlRrV1/Boa/USA/BC/2009) (Golden Gate virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Arenaviridae; Reptarenavirus; Golden reptarenavirus
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
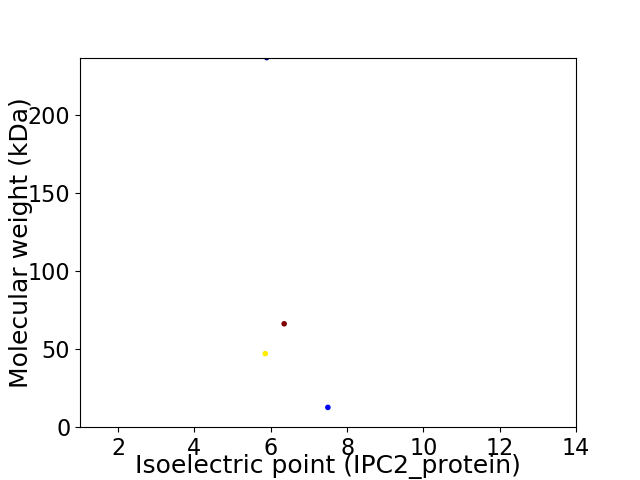
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|J7HBH4|GLYC_GOGV Glycoprotein OS=Alethinophid 1 reptarenavirus (isolate AlRrV1/Boa/USA/BC/2009) OX=1223562 GN=GPC PE=3 SV=1
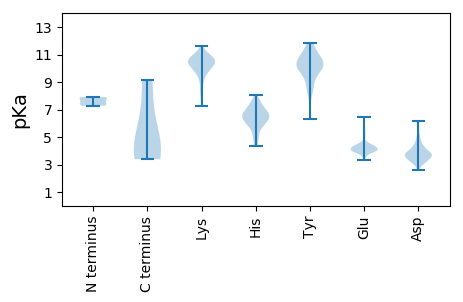
MM1 pKa = 7.28MSHH4 pKa = 6.62IRR6 pKa = 11.84LVLCSALLSMMSCPTSGTIGEE27 pKa = 4.46MLSLAASSNSSICHH41 pKa = 5.44GMQFTEE47 pKa = 4.24PTEE50 pKa = 4.25SFMGILPNEE59 pKa = 4.31TLPMFIFSIMHH70 pKa = 5.82SEE72 pKa = 4.09AVPKK76 pKa = 10.31VGKK79 pKa = 7.24TLRR82 pKa = 11.84ISHH85 pKa = 6.51DD86 pKa = 3.16MKK88 pKa = 11.12LFGGEE93 pKa = 3.91AVNYY97 pKa = 8.39MIFVNKK103 pKa = 9.91ILYY106 pKa = 9.87EE107 pKa = 4.43PISYY111 pKa = 10.83YY112 pKa = 10.95NYY114 pKa = 9.89TGSCRR119 pKa = 11.84QQGLSSCVRR128 pKa = 11.84VYY130 pKa = 11.48SDD132 pKa = 4.57LVNGSKK138 pKa = 10.89GNPSVKK144 pKa = 10.4LGFTAIEE151 pKa = 4.01LNKK154 pKa = 9.87TKK156 pKa = 9.72NTDD159 pKa = 3.77FHH161 pKa = 7.64NLGFKK166 pKa = 9.89ICFSCRR172 pKa = 11.84DD173 pKa = 3.51HH174 pKa = 8.05NGIRR178 pKa = 11.84LVVYY182 pKa = 10.09NKK184 pKa = 10.38NNRR187 pKa = 11.84TAQLMMCPSEE197 pKa = 5.57LIFSQDD203 pKa = 3.0FTINSTSVNPSEE215 pKa = 4.32EE216 pKa = 3.96AAVPLLNVTGYY227 pKa = 8.69TCIALHH233 pKa = 6.12NKK235 pKa = 9.58NMLTHH240 pKa = 6.87HH241 pKa = 6.48SPALSSGSKK250 pKa = 10.1VDD252 pKa = 3.46NTLEE256 pKa = 4.37PGCDD260 pKa = 3.31SNVGLFGHH268 pKa = 5.82STGTDD273 pKa = 3.69YY274 pKa = 11.67GWGLANFFSAGITNSLQISQLEE296 pKa = 4.34HH297 pKa = 5.01VTDD300 pKa = 6.18AIACKK305 pKa = 9.7IAKK308 pKa = 8.02TSNYY312 pKa = 6.3TTTALFLLNKK322 pKa = 10.02EE323 pKa = 3.94EE324 pKa = 4.5GEE326 pKa = 4.16IRR328 pKa = 11.84DD329 pKa = 3.66HH330 pKa = 7.37VIEE333 pKa = 5.1HH334 pKa = 6.04EE335 pKa = 4.28VALNYY340 pKa = 10.71LLAHH344 pKa = 5.98QGGLCSVVKK353 pKa = 10.85GPMCCSDD360 pKa = 2.91IDD362 pKa = 3.73DD363 pKa = 3.98FRR365 pKa = 11.84RR366 pKa = 11.84NVSDD370 pKa = 4.15MIDD373 pKa = 3.43KK374 pKa = 10.52VHH376 pKa = 6.86EE377 pKa = 4.18EE378 pKa = 3.85MKK380 pKa = 10.51KK381 pKa = 10.43FYY383 pKa = 10.66HH384 pKa = 6.44EE385 pKa = 4.57PDD387 pKa = 3.34PFGGLGTWGFYY398 pKa = 9.2GTIFGHH404 pKa = 5.09VLQWIPIIIMVVVVCFVCSWVRR426 pKa = 11.84KK427 pKa = 9.13
MM1 pKa = 7.28MSHH4 pKa = 6.62IRR6 pKa = 11.84LVLCSALLSMMSCPTSGTIGEE27 pKa = 4.46MLSLAASSNSSICHH41 pKa = 5.44GMQFTEE47 pKa = 4.24PTEE50 pKa = 4.25SFMGILPNEE59 pKa = 4.31TLPMFIFSIMHH70 pKa = 5.82SEE72 pKa = 4.09AVPKK76 pKa = 10.31VGKK79 pKa = 7.24TLRR82 pKa = 11.84ISHH85 pKa = 6.51DD86 pKa = 3.16MKK88 pKa = 11.12LFGGEE93 pKa = 3.91AVNYY97 pKa = 8.39MIFVNKK103 pKa = 9.91ILYY106 pKa = 9.87EE107 pKa = 4.43PISYY111 pKa = 10.83YY112 pKa = 10.95NYY114 pKa = 9.89TGSCRR119 pKa = 11.84QQGLSSCVRR128 pKa = 11.84VYY130 pKa = 11.48SDD132 pKa = 4.57LVNGSKK138 pKa = 10.89GNPSVKK144 pKa = 10.4LGFTAIEE151 pKa = 4.01LNKK154 pKa = 9.87TKK156 pKa = 9.72NTDD159 pKa = 3.77FHH161 pKa = 7.64NLGFKK166 pKa = 9.89ICFSCRR172 pKa = 11.84DD173 pKa = 3.51HH174 pKa = 8.05NGIRR178 pKa = 11.84LVVYY182 pKa = 10.09NKK184 pKa = 10.38NNRR187 pKa = 11.84TAQLMMCPSEE197 pKa = 5.57LIFSQDD203 pKa = 3.0FTINSTSVNPSEE215 pKa = 4.32EE216 pKa = 3.96AAVPLLNVTGYY227 pKa = 8.69TCIALHH233 pKa = 6.12NKK235 pKa = 9.58NMLTHH240 pKa = 6.87HH241 pKa = 6.48SPALSSGSKK250 pKa = 10.1VDD252 pKa = 3.46NTLEE256 pKa = 4.37PGCDD260 pKa = 3.31SNVGLFGHH268 pKa = 5.82STGTDD273 pKa = 3.69YY274 pKa = 11.67GWGLANFFSAGITNSLQISQLEE296 pKa = 4.34HH297 pKa = 5.01VTDD300 pKa = 6.18AIACKK305 pKa = 9.7IAKK308 pKa = 8.02TSNYY312 pKa = 6.3TTTALFLLNKK322 pKa = 10.02EE323 pKa = 3.94EE324 pKa = 4.5GEE326 pKa = 4.16IRR328 pKa = 11.84DD329 pKa = 3.66HH330 pKa = 7.37VIEE333 pKa = 5.1HH334 pKa = 6.04EE335 pKa = 4.28VALNYY340 pKa = 10.71LLAHH344 pKa = 5.98QGGLCSVVKK353 pKa = 10.85GPMCCSDD360 pKa = 2.91IDD362 pKa = 3.73DD363 pKa = 3.98FRR365 pKa = 11.84RR366 pKa = 11.84NVSDD370 pKa = 4.15MIDD373 pKa = 3.43KK374 pKa = 10.52VHH376 pKa = 6.86EE377 pKa = 4.18EE378 pKa = 3.85MKK380 pKa = 10.51KK381 pKa = 10.43FYY383 pKa = 10.66HH384 pKa = 6.44EE385 pKa = 4.57PDD387 pKa = 3.34PFGGLGTWGFYY398 pKa = 9.2GTIFGHH404 pKa = 5.09VLQWIPIIIMVVVVCFVCSWVRR426 pKa = 11.84KK427 pKa = 9.13
Molecular weight: 47.18 kDa
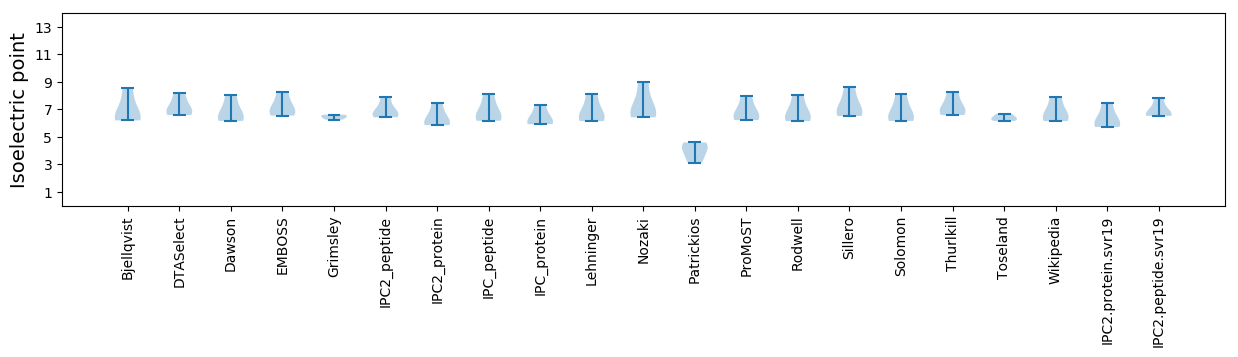
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|J7H5M4|NCAP_GOGV Nucleoprotein OS=Alethinophid 1 reptarenavirus (isolate AlRrV1/Boa/USA/BC/2009) OX=1223562 GN=N PE=3 SV=1
MM1 pKa = 7.4SGSTAIGLTTEE12 pKa = 4.4VISIITFILVIAIFVIEE29 pKa = 4.33IVSCVTMMTLKK40 pKa = 10.9AITLKK45 pKa = 10.64KK46 pKa = 10.26RR47 pKa = 11.84LSFCQGCGKK56 pKa = 9.8NASLVILPCKK66 pKa = 10.48NKK68 pKa = 9.76VCMEE72 pKa = 4.35CALKK76 pKa = 9.82MRR78 pKa = 11.84CPVCYY83 pKa = 9.02EE84 pKa = 3.65ACLWCEE90 pKa = 4.11NPDD93 pKa = 3.54GSLSSLALINKK104 pKa = 7.29EE105 pKa = 3.95RR106 pKa = 11.84NKK108 pKa = 9.9VRR110 pKa = 11.84DD111 pKa = 3.9NLPEE115 pKa = 3.89PP116 pKa = 4.2
MM1 pKa = 7.4SGSTAIGLTTEE12 pKa = 4.4VISIITFILVIAIFVIEE29 pKa = 4.33IVSCVTMMTLKK40 pKa = 10.9AITLKK45 pKa = 10.64KK46 pKa = 10.26RR47 pKa = 11.84LSFCQGCGKK56 pKa = 9.8NASLVILPCKK66 pKa = 10.48NKK68 pKa = 9.76VCMEE72 pKa = 4.35CALKK76 pKa = 9.82MRR78 pKa = 11.84CPVCYY83 pKa = 9.02EE84 pKa = 3.65ACLWCEE90 pKa = 4.11NPDD93 pKa = 3.54GSLSSLALINKK104 pKa = 7.29EE105 pKa = 3.95RR106 pKa = 11.84NKK108 pKa = 9.9VRR110 pKa = 11.84DD111 pKa = 3.9NLPEE115 pKa = 3.89PP116 pKa = 4.2
Molecular weight: 12.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3200 |
116 |
2066 |
800.0 |
90.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.219 ± 1.098 | 2.406 ± 0.852 |
5.813 ± 0.845 | 7.187 ± 0.734 |
4.719 ± 0.235 | 5.375 ± 0.496 |
1.875 ± 0.467 | 6.688 ± 0.424 |
7.75 ± 0.75 | 11.25 ± 1.219 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.094 ± 0.306 | 4.906 ± 0.317 |
4.281 ± 0.619 | 2.656 ± 0.299 |
4.656 ± 0.552 | 7.938 ± 0.472 |
5.594 ± 0.284 | 5.844 ± 0.372 |
1.0 ± 0.194 | 2.75 ± 0.4 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
