
Planococcus massiliensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Planococcaceae; Planococcus
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
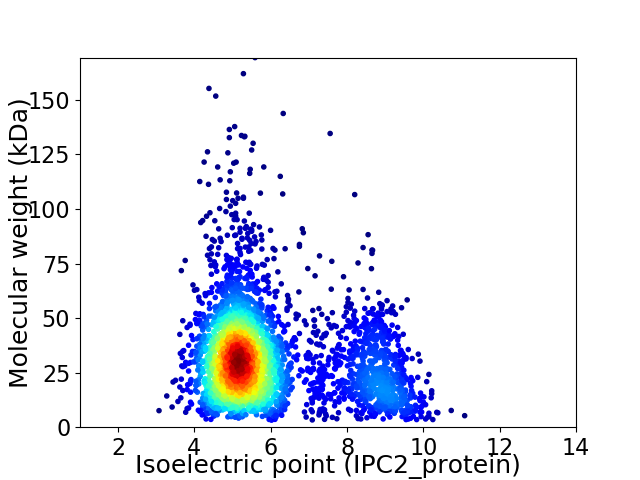
Virtual 2D-PAGE plot for 3353 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A098EI80|A0A098EI80_9BACL Gamma-DL-glutamyl hydrolase OS=Planococcus massiliensis OX=1499687 GN=pgdS PE=3 SV=1
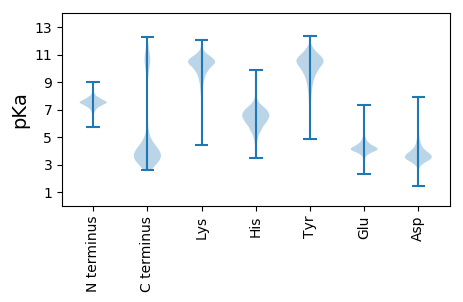
MM1 pKa = 7.72EE2 pKa = 5.48KK3 pKa = 10.69LNYY6 pKa = 10.34LILLLMAIFLVGCTDD21 pKa = 3.4IEE23 pKa = 4.3DD24 pKa = 3.93TASVEE29 pKa = 4.23EE30 pKa = 4.16TDD32 pKa = 3.41TDD34 pKa = 3.75VKK36 pKa = 10.74EE37 pKa = 4.16VSTVEE42 pKa = 4.11EE43 pKa = 4.33EE44 pKa = 4.39SSEE47 pKa = 4.21EE48 pKa = 3.86ATPEE52 pKa = 3.81AVEE55 pKa = 4.22EE56 pKa = 4.06EE57 pKa = 4.59LVAEE61 pKa = 4.21EE62 pKa = 4.64TPTEE66 pKa = 3.98EE67 pKa = 3.76TAAAPTDD74 pKa = 3.85DD75 pKa = 5.73LFAGYY80 pKa = 10.64NLIEE84 pKa = 4.21VDD86 pKa = 4.1GGDD89 pKa = 3.57LSGHH93 pKa = 6.18RR94 pKa = 11.84EE95 pKa = 3.97ANVVVDD101 pKa = 3.59IGFGDD106 pKa = 3.52RR107 pKa = 11.84EE108 pKa = 3.96YY109 pKa = 10.09WAFTNEE115 pKa = 3.57HH116 pKa = 5.5GQLVRR121 pKa = 11.84VIASEE126 pKa = 5.68IILQDD131 pKa = 4.01DD132 pKa = 3.44NTEE135 pKa = 4.19PVTSSGRR142 pKa = 11.84YY143 pKa = 8.54YY144 pKa = 9.99PDD146 pKa = 3.2EE147 pKa = 4.48AKK149 pKa = 10.97VPGVEE154 pKa = 5.25SPTLDD159 pKa = 3.7EE160 pKa = 4.32GHH162 pKa = 6.82VIADD166 pKa = 3.61SLGGVSNAYY175 pKa = 10.43NITPQEE181 pKa = 4.15STLNRR186 pKa = 11.84YY187 pKa = 8.77GDD189 pKa = 3.46QAYY192 pKa = 9.09MEE194 pKa = 4.31DD195 pKa = 4.39VIRR198 pKa = 11.84KK199 pKa = 9.54AGGAVDD205 pKa = 4.61FEE207 pKa = 5.36AIITYY212 pKa = 9.91PDD214 pKa = 3.66TEE216 pKa = 4.39TQIPSHH222 pKa = 5.38YY223 pKa = 10.19QYY225 pKa = 11.01TYY227 pKa = 9.12TVNGNKK233 pKa = 9.31VVDD236 pKa = 4.09NYY238 pKa = 11.79DD239 pKa = 3.3NVNPDD244 pKa = 3.55EE245 pKa = 4.5VNEE248 pKa = 4.18SLGLTGGDD256 pKa = 3.06APEE259 pKa = 4.15AAEE262 pKa = 4.89AEE264 pKa = 4.58PEE266 pKa = 4.27SAPAAPAPAAGGDD279 pKa = 3.76VSSVDD284 pKa = 3.28TDD286 pKa = 3.62GNGQVTIKK294 pKa = 9.56EE295 pKa = 3.92AKK297 pKa = 9.66AAGFSMPIMSDD308 pKa = 2.56HH309 pKa = 6.56WLYY312 pKa = 10.6PYY314 pKa = 10.18MRR316 pKa = 11.84DD317 pKa = 3.38NDD319 pKa = 3.64NDD321 pKa = 3.78GMVGEE326 pKa = 4.62
MM1 pKa = 7.72EE2 pKa = 5.48KK3 pKa = 10.69LNYY6 pKa = 10.34LILLLMAIFLVGCTDD21 pKa = 3.4IEE23 pKa = 4.3DD24 pKa = 3.93TASVEE29 pKa = 4.23EE30 pKa = 4.16TDD32 pKa = 3.41TDD34 pKa = 3.75VKK36 pKa = 10.74EE37 pKa = 4.16VSTVEE42 pKa = 4.11EE43 pKa = 4.33EE44 pKa = 4.39SSEE47 pKa = 4.21EE48 pKa = 3.86ATPEE52 pKa = 3.81AVEE55 pKa = 4.22EE56 pKa = 4.06EE57 pKa = 4.59LVAEE61 pKa = 4.21EE62 pKa = 4.64TPTEE66 pKa = 3.98EE67 pKa = 3.76TAAAPTDD74 pKa = 3.85DD75 pKa = 5.73LFAGYY80 pKa = 10.64NLIEE84 pKa = 4.21VDD86 pKa = 4.1GGDD89 pKa = 3.57LSGHH93 pKa = 6.18RR94 pKa = 11.84EE95 pKa = 3.97ANVVVDD101 pKa = 3.59IGFGDD106 pKa = 3.52RR107 pKa = 11.84EE108 pKa = 3.96YY109 pKa = 10.09WAFTNEE115 pKa = 3.57HH116 pKa = 5.5GQLVRR121 pKa = 11.84VIASEE126 pKa = 5.68IILQDD131 pKa = 4.01DD132 pKa = 3.44NTEE135 pKa = 4.19PVTSSGRR142 pKa = 11.84YY143 pKa = 8.54YY144 pKa = 9.99PDD146 pKa = 3.2EE147 pKa = 4.48AKK149 pKa = 10.97VPGVEE154 pKa = 5.25SPTLDD159 pKa = 3.7EE160 pKa = 4.32GHH162 pKa = 6.82VIADD166 pKa = 3.61SLGGVSNAYY175 pKa = 10.43NITPQEE181 pKa = 4.15STLNRR186 pKa = 11.84YY187 pKa = 8.77GDD189 pKa = 3.46QAYY192 pKa = 9.09MEE194 pKa = 4.31DD195 pKa = 4.39VIRR198 pKa = 11.84KK199 pKa = 9.54AGGAVDD205 pKa = 4.61FEE207 pKa = 5.36AIITYY212 pKa = 9.91PDD214 pKa = 3.66TEE216 pKa = 4.39TQIPSHH222 pKa = 5.38YY223 pKa = 10.19QYY225 pKa = 11.01TYY227 pKa = 9.12TVNGNKK233 pKa = 9.31VVDD236 pKa = 4.09NYY238 pKa = 11.79DD239 pKa = 3.3NVNPDD244 pKa = 3.55EE245 pKa = 4.5VNEE248 pKa = 4.18SLGLTGGDD256 pKa = 3.06APEE259 pKa = 4.15AAEE262 pKa = 4.89AEE264 pKa = 4.58PEE266 pKa = 4.27SAPAAPAPAAGGDD279 pKa = 3.76VSSVDD284 pKa = 3.28TDD286 pKa = 3.62GNGQVTIKK294 pKa = 9.56EE295 pKa = 3.92AKK297 pKa = 9.66AAGFSMPIMSDD308 pKa = 2.56HH309 pKa = 6.56WLYY312 pKa = 10.6PYY314 pKa = 10.18MRR316 pKa = 11.84DD317 pKa = 3.38NDD319 pKa = 3.64NDD321 pKa = 3.78GMVGEE326 pKa = 4.62
Molecular weight: 35.11 kDa
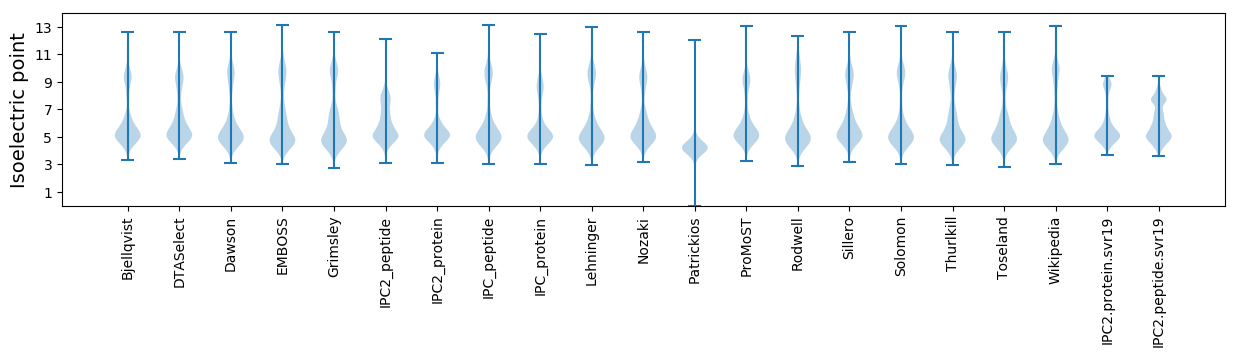
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A098EHE5|A0A098EHE5_9BACL Laccase domain protein OS=Planococcus massiliensis OX=1499687 GN=BN1080_00141 PE=3 SV=1
MM1 pKa = 7.61TLRR4 pKa = 11.84TYY6 pKa = 10.57QPNTRR11 pKa = 11.84KK12 pKa = 9.66HH13 pKa = 5.87SKK15 pKa = 8.83VHH17 pKa = 5.7GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.71NGRR29 pKa = 11.84RR30 pKa = 11.84VIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.8GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
MM1 pKa = 7.61TLRR4 pKa = 11.84TYY6 pKa = 10.57QPNTRR11 pKa = 11.84KK12 pKa = 9.66HH13 pKa = 5.87SKK15 pKa = 8.83VHH17 pKa = 5.7GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.71NGRR29 pKa = 11.84RR30 pKa = 11.84VIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.8GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
986772 |
29 |
1528 |
294.3 |
32.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.417 ± 0.046 | 0.556 ± 0.01 |
5.108 ± 0.03 | 8.054 ± 0.053 |
4.62 ± 0.036 | 7.168 ± 0.043 |
2.002 ± 0.021 | 7.258 ± 0.04 |
6.209 ± 0.035 | 9.8 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.937 ± 0.021 | 3.834 ± 0.031 |
3.764 ± 0.02 | 3.655 ± 0.03 |
4.166 ± 0.032 | 5.905 ± 0.037 |
5.274 ± 0.029 | 6.975 ± 0.035 |
1.029 ± 0.017 | 3.267 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
