
McMurdo Ice Shelf pond-associated circular DNA virus-3
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins
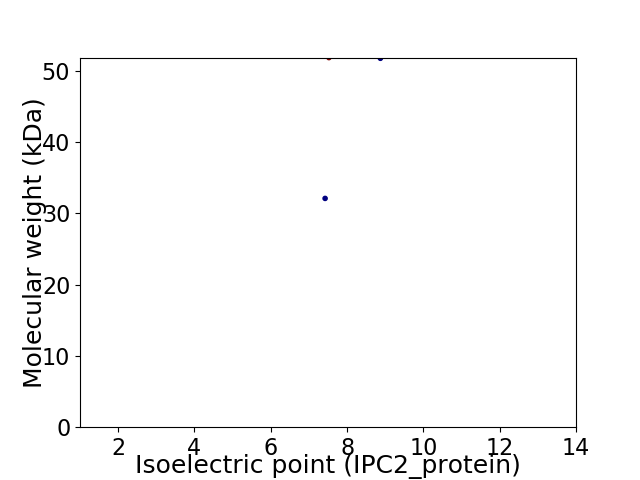
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
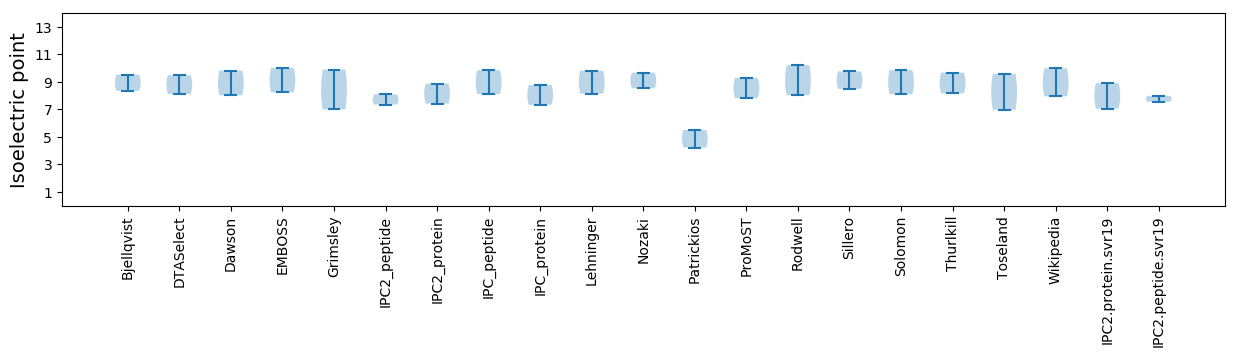
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075M3P7|A0A075M3P7_9VIRU Putative capsid protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-3 OX=1521387 PE=4 SV=1
MM1 pKa = 7.4SKK3 pKa = 10.19NRR5 pKa = 11.84NFVFTWNNYY14 pKa = 8.55SDD16 pKa = 3.72ASKK19 pKa = 9.8TYY21 pKa = 10.82LSTLACKK28 pKa = 9.67YY29 pKa = 9.19VAYY32 pKa = 10.13AEE34 pKa = 4.49EE35 pKa = 4.31VAPTTGTRR43 pKa = 11.84HH44 pKa = 5.11LQGFIAFTNAKK55 pKa = 8.95TIQQARR61 pKa = 11.84SKK63 pKa = 11.15LPGCHH68 pKa = 5.89VEE70 pKa = 4.29TMNGSIAQSEE80 pKa = 4.98DD81 pKa = 3.59YY82 pKa = 10.51CSKK85 pKa = 11.06AGTLTEE91 pKa = 5.1HH92 pKa = 5.6GTKK95 pKa = 10.26PISNDD100 pKa = 2.95NNGRR104 pKa = 11.84AEE106 pKa = 3.91KK107 pKa = 10.65LRR109 pKa = 11.84WQRR112 pKa = 11.84ARR114 pKa = 11.84DD115 pKa = 3.52FAKK118 pKa = 10.36EE119 pKa = 3.97GKK121 pKa = 9.93LDD123 pKa = 4.8EE124 pKa = 5.0IDD126 pKa = 3.3ADD128 pKa = 3.65IFIRR132 pKa = 11.84CYY134 pKa = 9.92STLKK138 pKa = 10.39RR139 pKa = 11.84IKK141 pKa = 10.01SDD143 pKa = 3.5YY144 pKa = 8.51ATKK147 pKa = 9.54PQPIDD152 pKa = 3.61PVCIWIYY159 pKa = 11.03GPTGTGKK166 pKa = 9.45SHH168 pKa = 6.59AVEE171 pKa = 4.22TRR173 pKa = 11.84FPNCYY178 pKa = 9.04KK179 pKa = 10.8KK180 pKa = 11.11CMDD183 pKa = 4.11DD184 pKa = 4.98LKK186 pKa = 10.92WFDD189 pKa = 4.09GYY191 pKa = 11.52AHH193 pKa = 7.43EE194 pKa = 4.76DD195 pKa = 3.41AIYY198 pKa = 10.97LEE200 pKa = 6.04DD201 pKa = 3.52IDD203 pKa = 5.02KK204 pKa = 11.27YY205 pKa = 7.36QVKK208 pKa = 9.58WGGILKK214 pKa = 10.22RR215 pKa = 11.84LADD218 pKa = 4.59RR219 pKa = 11.84WPMQASIKK227 pKa = 10.64GAMAYY232 pKa = 9.19IRR234 pKa = 11.84PKK236 pKa = 10.48FVLVTSNYY244 pKa = 10.19RR245 pKa = 11.84IDD247 pKa = 5.29EE248 pKa = 4.17IWTDD252 pKa = 3.79PQTLEE257 pKa = 3.55PLQRR261 pKa = 11.84RR262 pKa = 11.84FTEE265 pKa = 4.13IEE267 pKa = 4.16KK268 pKa = 9.18LTQEE272 pKa = 3.96QVIDD276 pKa = 4.19FDD278 pKa = 3.77QQ279 pKa = 4.78
MM1 pKa = 7.4SKK3 pKa = 10.19NRR5 pKa = 11.84NFVFTWNNYY14 pKa = 8.55SDD16 pKa = 3.72ASKK19 pKa = 9.8TYY21 pKa = 10.82LSTLACKK28 pKa = 9.67YY29 pKa = 9.19VAYY32 pKa = 10.13AEE34 pKa = 4.49EE35 pKa = 4.31VAPTTGTRR43 pKa = 11.84HH44 pKa = 5.11LQGFIAFTNAKK55 pKa = 8.95TIQQARR61 pKa = 11.84SKK63 pKa = 11.15LPGCHH68 pKa = 5.89VEE70 pKa = 4.29TMNGSIAQSEE80 pKa = 4.98DD81 pKa = 3.59YY82 pKa = 10.51CSKK85 pKa = 11.06AGTLTEE91 pKa = 5.1HH92 pKa = 5.6GTKK95 pKa = 10.26PISNDD100 pKa = 2.95NNGRR104 pKa = 11.84AEE106 pKa = 3.91KK107 pKa = 10.65LRR109 pKa = 11.84WQRR112 pKa = 11.84ARR114 pKa = 11.84DD115 pKa = 3.52FAKK118 pKa = 10.36EE119 pKa = 3.97GKK121 pKa = 9.93LDD123 pKa = 4.8EE124 pKa = 5.0IDD126 pKa = 3.3ADD128 pKa = 3.65IFIRR132 pKa = 11.84CYY134 pKa = 9.92STLKK138 pKa = 10.39RR139 pKa = 11.84IKK141 pKa = 10.01SDD143 pKa = 3.5YY144 pKa = 8.51ATKK147 pKa = 9.54PQPIDD152 pKa = 3.61PVCIWIYY159 pKa = 11.03GPTGTGKK166 pKa = 9.45SHH168 pKa = 6.59AVEE171 pKa = 4.22TRR173 pKa = 11.84FPNCYY178 pKa = 9.04KK179 pKa = 10.8KK180 pKa = 11.11CMDD183 pKa = 4.11DD184 pKa = 4.98LKK186 pKa = 10.92WFDD189 pKa = 4.09GYY191 pKa = 11.52AHH193 pKa = 7.43EE194 pKa = 4.76DD195 pKa = 3.41AIYY198 pKa = 10.97LEE200 pKa = 6.04DD201 pKa = 3.52IDD203 pKa = 5.02KK204 pKa = 11.27YY205 pKa = 7.36QVKK208 pKa = 9.58WGGILKK214 pKa = 10.22RR215 pKa = 11.84LADD218 pKa = 4.59RR219 pKa = 11.84WPMQASIKK227 pKa = 10.64GAMAYY232 pKa = 9.19IRR234 pKa = 11.84PKK236 pKa = 10.48FVLVTSNYY244 pKa = 10.19RR245 pKa = 11.84IDD247 pKa = 5.29EE248 pKa = 4.17IWTDD252 pKa = 3.79PQTLEE257 pKa = 3.55PLQRR261 pKa = 11.84RR262 pKa = 11.84FTEE265 pKa = 4.13IEE267 pKa = 4.16KK268 pKa = 9.18LTQEE272 pKa = 3.96QVIDD276 pKa = 4.19FDD278 pKa = 3.77QQ279 pKa = 4.78
Molecular weight: 32.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075M3P7|A0A075M3P7_9VIRU Putative capsid protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-3 OX=1521387 PE=4 SV=1
MM1 pKa = 7.99PNRR4 pKa = 11.84KK5 pKa = 8.46VVKK8 pKa = 9.24KK9 pKa = 10.08SKK11 pKa = 10.03RR12 pKa = 11.84KK13 pKa = 8.46RR14 pKa = 11.84TFGQKK19 pKa = 9.03GLRR22 pKa = 11.84MLSQAAGAAVGYY34 pKa = 7.6GTHH37 pKa = 6.42NLAGIAGGAEE47 pKa = 4.0VGGYY51 pKa = 10.38LYY53 pKa = 11.17DD54 pKa = 3.81KK55 pKa = 10.24MLGEE59 pKa = 5.28AYY61 pKa = 10.28DD62 pKa = 3.91DD63 pKa = 4.1LGRR66 pKa = 11.84DD67 pKa = 3.54AMVEE71 pKa = 4.0DD72 pKa = 5.05AVNDD76 pKa = 3.92LNRR79 pKa = 11.84AEE81 pKa = 4.09AADD84 pKa = 3.79RR85 pKa = 11.84FDD87 pKa = 3.8RR88 pKa = 11.84KK89 pKa = 10.02KK90 pKa = 10.65DD91 pKa = 3.49SSMGAYY97 pKa = 9.63AGKK100 pKa = 10.04FKK102 pKa = 11.02KK103 pKa = 9.9NRR105 pKa = 11.84RR106 pKa = 11.84LKK108 pKa = 10.69KK109 pKa = 10.42SSLKK113 pKa = 10.76LKK115 pKa = 10.34ALSNGYY121 pKa = 9.11HH122 pKa = 5.76ATSEE126 pKa = 4.12HH127 pKa = 6.56FGRR130 pKa = 11.84IEE132 pKa = 4.92DD133 pKa = 4.06PNCVYY138 pKa = 10.32IHH140 pKa = 7.1HH141 pKa = 6.43STFDD145 pKa = 3.77NIQSIRR151 pKa = 11.84AISGAMLRR159 pKa = 11.84KK160 pKa = 9.59LFKK163 pKa = 10.41QAGIPLTDD171 pKa = 3.75KK172 pKa = 11.27NEE174 pKa = 4.51EE175 pKa = 3.96IPASNWEE182 pKa = 4.2SSAGTKK188 pKa = 9.28LQYY191 pKa = 10.82VHH193 pKa = 7.34LDD195 pKa = 3.49QMSGAQTEE203 pKa = 3.54IDD205 pKa = 3.73YY206 pKa = 10.93IVTNNQGLSQILANWPAMQNHH227 pKa = 6.43ILSYY231 pKa = 10.94LNGIAVTVPWKK242 pKa = 10.67LCLFIPDD249 pKa = 4.17VGPVITTHH257 pKa = 6.73RR258 pKa = 11.84CLSQISLWSEE268 pKa = 3.71HH269 pKa = 5.19LTVFQSSEE277 pKa = 3.5IKK279 pKa = 9.72VQNRR283 pKa = 11.84TKK285 pKa = 11.18GDD287 pKa = 3.56LSGVADD293 pKa = 4.39PEE295 pKa = 3.82AAAIEE300 pKa = 4.43RR301 pKa = 11.84VDD303 pKa = 3.93SQPLIARR310 pKa = 11.84TYY312 pKa = 8.03YY313 pKa = 10.64LKK315 pKa = 11.04SGDD318 pKa = 2.93IRR320 pKa = 11.84LRR322 pKa = 11.84SNAGGLANSNNVFTAAAVTGLNLIRR347 pKa = 11.84SAQLADD353 pKa = 3.54ISWQNRR359 pKa = 11.84PVPQIFSTCSKK370 pKa = 10.63SADD373 pKa = 3.83TILQPGDD380 pKa = 3.05IKK382 pKa = 10.72KK383 pKa = 8.44TFVAHH388 pKa = 6.63KK389 pKa = 10.22FSGLFLNVLIKK400 pKa = 10.01MRR402 pKa = 11.84AHH404 pKa = 6.79QINAPGTFANGCQGRR419 pKa = 11.84SEE421 pKa = 4.18LMCFEE426 pKa = 4.25EE427 pKa = 5.38KK428 pKa = 10.3IRR430 pKa = 11.84TAGTNKK436 pKa = 9.66IVIQYY441 pKa = 6.79EE442 pKa = 3.66QHH444 pKa = 5.29YY445 pKa = 9.58QAGAFLKK452 pKa = 10.48SGKK455 pKa = 9.34KK456 pKa = 9.72VPFISTLTVTEE467 pKa = 4.29KK468 pKa = 11.35NNVV471 pKa = 3.13
MM1 pKa = 7.99PNRR4 pKa = 11.84KK5 pKa = 8.46VVKK8 pKa = 9.24KK9 pKa = 10.08SKK11 pKa = 10.03RR12 pKa = 11.84KK13 pKa = 8.46RR14 pKa = 11.84TFGQKK19 pKa = 9.03GLRR22 pKa = 11.84MLSQAAGAAVGYY34 pKa = 7.6GTHH37 pKa = 6.42NLAGIAGGAEE47 pKa = 4.0VGGYY51 pKa = 10.38LYY53 pKa = 11.17DD54 pKa = 3.81KK55 pKa = 10.24MLGEE59 pKa = 5.28AYY61 pKa = 10.28DD62 pKa = 3.91DD63 pKa = 4.1LGRR66 pKa = 11.84DD67 pKa = 3.54AMVEE71 pKa = 4.0DD72 pKa = 5.05AVNDD76 pKa = 3.92LNRR79 pKa = 11.84AEE81 pKa = 4.09AADD84 pKa = 3.79RR85 pKa = 11.84FDD87 pKa = 3.8RR88 pKa = 11.84KK89 pKa = 10.02KK90 pKa = 10.65DD91 pKa = 3.49SSMGAYY97 pKa = 9.63AGKK100 pKa = 10.04FKK102 pKa = 11.02KK103 pKa = 9.9NRR105 pKa = 11.84RR106 pKa = 11.84LKK108 pKa = 10.69KK109 pKa = 10.42SSLKK113 pKa = 10.76LKK115 pKa = 10.34ALSNGYY121 pKa = 9.11HH122 pKa = 5.76ATSEE126 pKa = 4.12HH127 pKa = 6.56FGRR130 pKa = 11.84IEE132 pKa = 4.92DD133 pKa = 4.06PNCVYY138 pKa = 10.32IHH140 pKa = 7.1HH141 pKa = 6.43STFDD145 pKa = 3.77NIQSIRR151 pKa = 11.84AISGAMLRR159 pKa = 11.84KK160 pKa = 9.59LFKK163 pKa = 10.41QAGIPLTDD171 pKa = 3.75KK172 pKa = 11.27NEE174 pKa = 4.51EE175 pKa = 3.96IPASNWEE182 pKa = 4.2SSAGTKK188 pKa = 9.28LQYY191 pKa = 10.82VHH193 pKa = 7.34LDD195 pKa = 3.49QMSGAQTEE203 pKa = 3.54IDD205 pKa = 3.73YY206 pKa = 10.93IVTNNQGLSQILANWPAMQNHH227 pKa = 6.43ILSYY231 pKa = 10.94LNGIAVTVPWKK242 pKa = 10.67LCLFIPDD249 pKa = 4.17VGPVITTHH257 pKa = 6.73RR258 pKa = 11.84CLSQISLWSEE268 pKa = 3.71HH269 pKa = 5.19LTVFQSSEE277 pKa = 3.5IKK279 pKa = 9.72VQNRR283 pKa = 11.84TKK285 pKa = 11.18GDD287 pKa = 3.56LSGVADD293 pKa = 4.39PEE295 pKa = 3.82AAAIEE300 pKa = 4.43RR301 pKa = 11.84VDD303 pKa = 3.93SQPLIARR310 pKa = 11.84TYY312 pKa = 8.03YY313 pKa = 10.64LKK315 pKa = 11.04SGDD318 pKa = 2.93IRR320 pKa = 11.84LRR322 pKa = 11.84SNAGGLANSNNVFTAAAVTGLNLIRR347 pKa = 11.84SAQLADD353 pKa = 3.54ISWQNRR359 pKa = 11.84PVPQIFSTCSKK370 pKa = 10.63SADD373 pKa = 3.83TILQPGDD380 pKa = 3.05IKK382 pKa = 10.72KK383 pKa = 8.44TFVAHH388 pKa = 6.63KK389 pKa = 10.22FSGLFLNVLIKK400 pKa = 10.01MRR402 pKa = 11.84AHH404 pKa = 6.79QINAPGTFANGCQGRR419 pKa = 11.84SEE421 pKa = 4.18LMCFEE426 pKa = 4.25EE427 pKa = 5.38KK428 pKa = 10.3IRR430 pKa = 11.84TAGTNKK436 pKa = 9.66IVIQYY441 pKa = 6.79EE442 pKa = 3.66QHH444 pKa = 5.29YY445 pKa = 9.58QAGAFLKK452 pKa = 10.48SGKK455 pKa = 9.34KK456 pKa = 9.72VPFISTLTVTEE467 pKa = 4.29KK468 pKa = 11.35NNVV471 pKa = 3.13
Molecular weight: 51.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
750 |
279 |
471 |
375.0 |
41.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.067 ± 0.665 | 1.733 ± 0.437 |
5.6 ± 0.883 | 4.667 ± 0.602 |
3.733 ± 0.118 | 6.933 ± 0.877 |
2.267 ± 0.267 | 6.8 ± 0.208 |
7.733 ± 0.287 | 7.467 ± 0.975 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.0 ± 0.117 | 5.2 ± 0.708 |
3.6 ± 0.395 | 4.8 ± 0.079 |
5.333 ± 0.226 | 6.667 ± 0.929 |
6.267 ± 0.912 | 4.8 ± 0.685 |
1.6 ± 0.512 | 3.733 ± 0.724 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
