
Furculomyces boomerangus
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Zoopagomycota; Kickxellomycotina; Harpellomycetes; Harpellales; Harpellaceae; Furculomyces
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
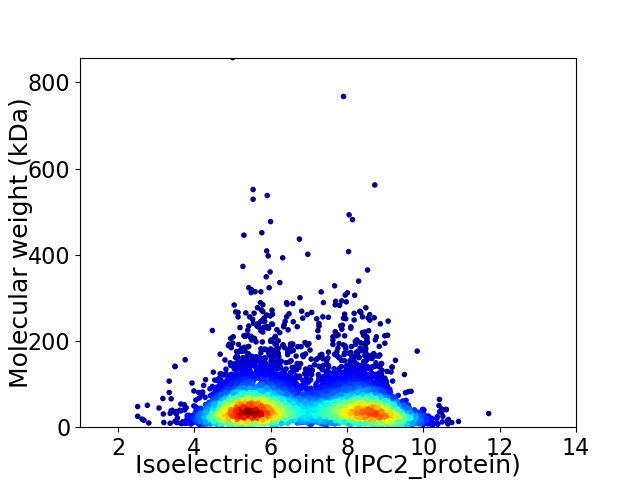
Virtual 2D-PAGE plot for 6645 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2T9Z5P6|A0A2T9Z5P6_9FUNG 2Fe-2S ferredoxin-type domain-containing protein OS=Furculomyces boomerangus OX=61424 GN=BB559_000328 PE=4 SV=1
MM1 pKa = 7.12QCPFSSYY8 pKa = 10.33CKK10 pKa = 10.31KK11 pKa = 10.21VDD13 pKa = 3.51NMLISCIPIIPKK25 pKa = 8.05TTVSVPSTTTFLPTLTSDD43 pKa = 3.22SSLATTSLAAEE54 pKa = 4.39TGSGQEE60 pKa = 3.82PTQIDD65 pKa = 3.8VNATSTGALSTDD77 pKa = 3.03SDD79 pKa = 3.76VTIIVTEE86 pKa = 4.09PPPTITEE93 pKa = 4.02TDD95 pKa = 3.34QSVVTLFTT103 pKa = 4.03
MM1 pKa = 7.12QCPFSSYY8 pKa = 10.33CKK10 pKa = 10.31KK11 pKa = 10.21VDD13 pKa = 3.51NMLISCIPIIPKK25 pKa = 8.05TTVSVPSTTTFLPTLTSDD43 pKa = 3.22SSLATTSLAAEE54 pKa = 4.39TGSGQEE60 pKa = 3.82PTQIDD65 pKa = 3.8VNATSTGALSTDD77 pKa = 3.03SDD79 pKa = 3.76VTIIVTEE86 pKa = 4.09PPPTITEE93 pKa = 4.02TDD95 pKa = 3.34QSVVTLFTT103 pKa = 4.03
Molecular weight: 10.76 kDa
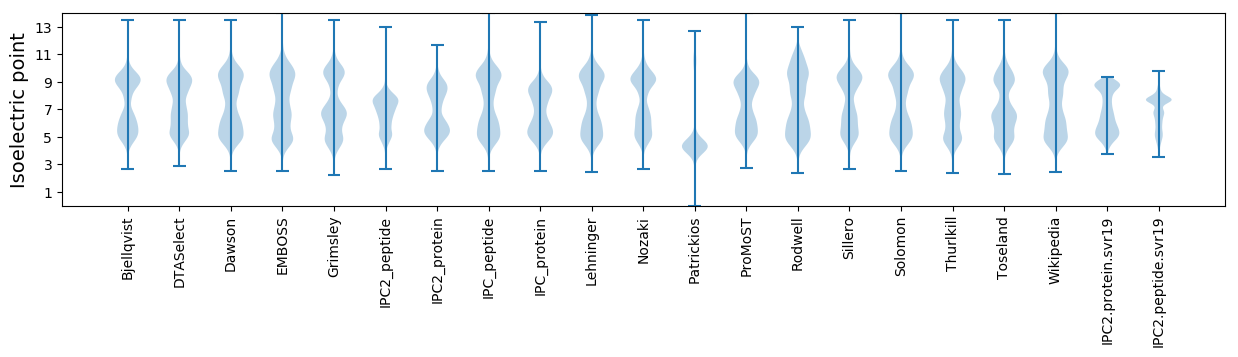
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T9YBV1|A0A2T9YBV1_9FUNG Uncharacterized protein OS=Furculomyces boomerangus OX=61424 GN=BB559_002007 PE=3 SV=1
MM1 pKa = 7.71RR2 pKa = 11.84SSSSSLSLSQSLNSSSSQLRR22 pKa = 11.84SSSSSQLRR30 pKa = 11.84SSSSSILSSQPTMSSNSPSFSQSVRR55 pKa = 11.84SSSIPASSQPPRR67 pKa = 11.84PSNSPTSSQPPRR79 pKa = 11.84PSNSPTFSQPPRR91 pKa = 11.84PSNSPTFSQPPRR103 pKa = 11.84PSNSPTSNQPPRR115 pKa = 11.84PSTTPASSQPPRR127 pKa = 11.84PSTTPTSSQPPRR139 pKa = 11.84PSNSPTSNQPPRR151 pKa = 11.84PSTTPASSQPPRR163 pKa = 11.84PSTTPASSQPLRR175 pKa = 11.84PSNNPTSNQPPRR187 pKa = 11.84PSNNPTSSQPSRR199 pKa = 11.84PSNNPTSSQPARR211 pKa = 11.84PSSSSPPRR219 pKa = 11.84QSTRR223 pKa = 11.84PPSNSPPSQPARR235 pKa = 11.84PSSSSPPRR243 pKa = 11.84QSSRR247 pKa = 11.84PPSNSPPRR255 pKa = 11.84QSSRR259 pKa = 11.84PPSNSPPRR267 pKa = 11.84QSSRR271 pKa = 11.84PPSNSPPRR279 pKa = 11.84QSSRR283 pKa = 11.84PPSNSPPRR291 pKa = 11.84QSSRR295 pKa = 11.84PPSNSSRR302 pKa = 11.84RR303 pKa = 3.68
MM1 pKa = 7.71RR2 pKa = 11.84SSSSSLSLSQSLNSSSSQLRR22 pKa = 11.84SSSSSQLRR30 pKa = 11.84SSSSSILSSQPTMSSNSPSFSQSVRR55 pKa = 11.84SSSIPASSQPPRR67 pKa = 11.84PSNSPTSSQPPRR79 pKa = 11.84PSNSPTFSQPPRR91 pKa = 11.84PSNSPTFSQPPRR103 pKa = 11.84PSNSPTSNQPPRR115 pKa = 11.84PSTTPASSQPPRR127 pKa = 11.84PSTTPTSSQPPRR139 pKa = 11.84PSNSPTSNQPPRR151 pKa = 11.84PSTTPASSQPPRR163 pKa = 11.84PSTTPASSQPLRR175 pKa = 11.84PSNNPTSNQPPRR187 pKa = 11.84PSNNPTSSQPSRR199 pKa = 11.84PSNNPTSSQPARR211 pKa = 11.84PSSSSPPRR219 pKa = 11.84QSTRR223 pKa = 11.84PPSNSPPSQPARR235 pKa = 11.84PSSSSPPRR243 pKa = 11.84QSSRR247 pKa = 11.84PPSNSPPRR255 pKa = 11.84QSSRR259 pKa = 11.84PPSNSPPRR267 pKa = 11.84QSSRR271 pKa = 11.84PPSNSPPRR279 pKa = 11.84QSSRR283 pKa = 11.84PPSNSPPRR291 pKa = 11.84QSSRR295 pKa = 11.84PPSNSSRR302 pKa = 11.84RR303 pKa = 3.68
Molecular weight: 31.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3304190 |
50 |
7614 |
497.2 |
56.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.723 ± 0.026 | 1.174 ± 0.012 |
5.362 ± 0.02 | 6.513 ± 0.028 |
4.366 ± 0.019 | 5.235 ± 0.027 |
1.898 ± 0.012 | 7.18 ± 0.025 |
7.974 ± 0.033 | 8.802 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.075 ± 0.01 | 7.291 ± 0.034 |
4.272 ± 0.024 | 3.797 ± 0.018 |
3.885 ± 0.018 | 9.739 ± 0.043 |
5.847 ± 0.024 | 5.475 ± 0.023 |
0.865 ± 0.008 | 3.526 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
