
Desulfovibrio senegalensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
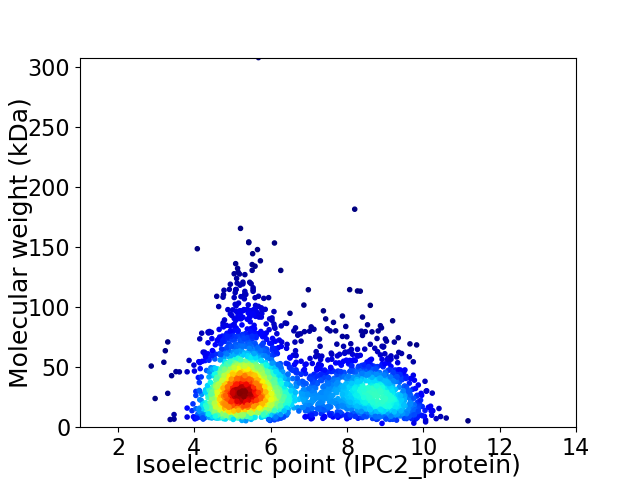
Virtual 2D-PAGE plot for 3062 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N6N609|A0A6N6N609_9DELT TrkA family potassium uptake protein OS=Desulfovibrio senegalensis OX=1721087 GN=F8A88_04455 PE=4 SV=1
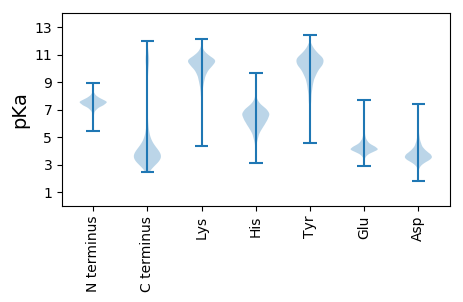
MM1 pKa = 7.3KK2 pKa = 10.1RR3 pKa = 11.84LVMLAVLCAFAFGMAATAASAADD26 pKa = 3.39IKK28 pKa = 11.05ATGTYY33 pKa = 8.68TFEE36 pKa = 4.86AVWGDD41 pKa = 3.48NSFDD45 pKa = 6.22DD46 pKa = 5.76DD47 pKa = 4.96DD48 pKa = 4.4QDD50 pKa = 4.39SDD52 pKa = 3.77FDD54 pKa = 4.07VYY56 pKa = 11.17QRR58 pKa = 11.84LRR60 pKa = 11.84TKK62 pKa = 10.76FEE64 pKa = 4.61FIANEE69 pKa = 3.79NLKK72 pKa = 10.57GVLYY76 pKa = 10.13TEE78 pKa = 4.99VGTSTWGSTMHH89 pKa = 6.4VGTDD93 pKa = 2.89IDD95 pKa = 4.07DD96 pKa = 4.2LVTIKK101 pKa = 10.75AGYY104 pKa = 9.78IDD106 pKa = 5.02FNWPGTQQNIKK117 pKa = 9.83VGHH120 pKa = 6.42FGVALPNAVGGSIILDD136 pKa = 3.87DD137 pKa = 4.68EE138 pKa = 4.79LPSAVITGPITDD150 pKa = 3.59NVSYY154 pKa = 11.12LLGWHH159 pKa = 6.81RR160 pKa = 11.84LDD162 pKa = 5.24KK163 pKa = 11.24SGAQNGTDD171 pKa = 3.73DD172 pKa = 4.4EE173 pKa = 5.47LDD175 pKa = 3.62AVAMALPLNFDD186 pKa = 4.06GVSVAPFILAAYY198 pKa = 10.09AGDD201 pKa = 3.97NYY203 pKa = 10.8DD204 pKa = 4.05ANVTNSITWAGVSFEE219 pKa = 4.65LTMFDD224 pKa = 3.55PFVFKK229 pKa = 11.05ADD231 pKa = 3.92LNYY234 pKa = 10.73GQADD238 pKa = 3.82YY239 pKa = 11.5EE240 pKa = 4.21NAEE243 pKa = 4.76DD244 pKa = 3.73QDD246 pKa = 3.21GWYY249 pKa = 9.93FATSAEE255 pKa = 4.2YY256 pKa = 10.47KK257 pKa = 10.58GFDD260 pKa = 3.57FMTPEE265 pKa = 3.78AFFVYY270 pKa = 8.09TTGDD274 pKa = 3.88DD275 pKa = 3.87NNVTYY280 pKa = 10.58AGEE283 pKa = 4.12DD284 pKa = 3.63QMPLLAGDD292 pKa = 3.64WAVGSFFFGGDD303 pKa = 2.91WGIGAQSSIDD313 pKa = 4.3ADD315 pKa = 4.27ANWLGFWTLGVSLKK329 pKa = 10.68DD330 pKa = 3.03ISFFEE335 pKa = 4.64GLTHH339 pKa = 6.17TVNVLYY345 pKa = 10.32IQGTNDD351 pKa = 3.29KK352 pKa = 10.76DD353 pKa = 3.53NFIAGDD359 pKa = 3.73LTYY362 pKa = 11.11LNEE365 pKa = 5.87DD366 pKa = 3.49DD367 pKa = 5.68SLWEE371 pKa = 4.23LDD373 pKa = 3.97LNSAYY378 pKa = 10.56SIYY381 pKa = 10.71DD382 pKa = 3.27EE383 pKa = 3.96LTAYY387 pKa = 10.66VEE389 pKa = 4.69LGWIAPDD396 pKa = 3.32WDD398 pKa = 3.54NDD400 pKa = 3.54RR401 pKa = 11.84NINDD405 pKa = 3.47DD406 pKa = 4.03EE407 pKa = 4.2AWKK410 pKa = 10.95VSTGLNYY417 pKa = 9.91VFF419 pKa = 5.39
MM1 pKa = 7.3KK2 pKa = 10.1RR3 pKa = 11.84LVMLAVLCAFAFGMAATAASAADD26 pKa = 3.39IKK28 pKa = 11.05ATGTYY33 pKa = 8.68TFEE36 pKa = 4.86AVWGDD41 pKa = 3.48NSFDD45 pKa = 6.22DD46 pKa = 5.76DD47 pKa = 4.96DD48 pKa = 4.4QDD50 pKa = 4.39SDD52 pKa = 3.77FDD54 pKa = 4.07VYY56 pKa = 11.17QRR58 pKa = 11.84LRR60 pKa = 11.84TKK62 pKa = 10.76FEE64 pKa = 4.61FIANEE69 pKa = 3.79NLKK72 pKa = 10.57GVLYY76 pKa = 10.13TEE78 pKa = 4.99VGTSTWGSTMHH89 pKa = 6.4VGTDD93 pKa = 2.89IDD95 pKa = 4.07DD96 pKa = 4.2LVTIKK101 pKa = 10.75AGYY104 pKa = 9.78IDD106 pKa = 5.02FNWPGTQQNIKK117 pKa = 9.83VGHH120 pKa = 6.42FGVALPNAVGGSIILDD136 pKa = 3.87DD137 pKa = 4.68EE138 pKa = 4.79LPSAVITGPITDD150 pKa = 3.59NVSYY154 pKa = 11.12LLGWHH159 pKa = 6.81RR160 pKa = 11.84LDD162 pKa = 5.24KK163 pKa = 11.24SGAQNGTDD171 pKa = 3.73DD172 pKa = 4.4EE173 pKa = 5.47LDD175 pKa = 3.62AVAMALPLNFDD186 pKa = 4.06GVSVAPFILAAYY198 pKa = 10.09AGDD201 pKa = 3.97NYY203 pKa = 10.8DD204 pKa = 4.05ANVTNSITWAGVSFEE219 pKa = 4.65LTMFDD224 pKa = 3.55PFVFKK229 pKa = 11.05ADD231 pKa = 3.92LNYY234 pKa = 10.73GQADD238 pKa = 3.82YY239 pKa = 11.5EE240 pKa = 4.21NAEE243 pKa = 4.76DD244 pKa = 3.73QDD246 pKa = 3.21GWYY249 pKa = 9.93FATSAEE255 pKa = 4.2YY256 pKa = 10.47KK257 pKa = 10.58GFDD260 pKa = 3.57FMTPEE265 pKa = 3.78AFFVYY270 pKa = 8.09TTGDD274 pKa = 3.88DD275 pKa = 3.87NNVTYY280 pKa = 10.58AGEE283 pKa = 4.12DD284 pKa = 3.63QMPLLAGDD292 pKa = 3.64WAVGSFFFGGDD303 pKa = 2.91WGIGAQSSIDD313 pKa = 4.3ADD315 pKa = 4.27ANWLGFWTLGVSLKK329 pKa = 10.68DD330 pKa = 3.03ISFFEE335 pKa = 4.64GLTHH339 pKa = 6.17TVNVLYY345 pKa = 10.32IQGTNDD351 pKa = 3.29KK352 pKa = 10.76DD353 pKa = 3.53NFIAGDD359 pKa = 3.73LTYY362 pKa = 11.11LNEE365 pKa = 5.87DD366 pKa = 3.49DD367 pKa = 5.68SLWEE371 pKa = 4.23LDD373 pKa = 3.97LNSAYY378 pKa = 10.56SIYY381 pKa = 10.71DD382 pKa = 3.27EE383 pKa = 3.96LTAYY387 pKa = 10.66VEE389 pKa = 4.69LGWIAPDD396 pKa = 3.32WDD398 pKa = 3.54NDD400 pKa = 3.54RR401 pKa = 11.84NINDD405 pKa = 3.47DD406 pKa = 4.03EE407 pKa = 4.2AWKK410 pKa = 10.95VSTGLNYY417 pKa = 9.91VFF419 pKa = 5.39
Molecular weight: 46.11 kDa
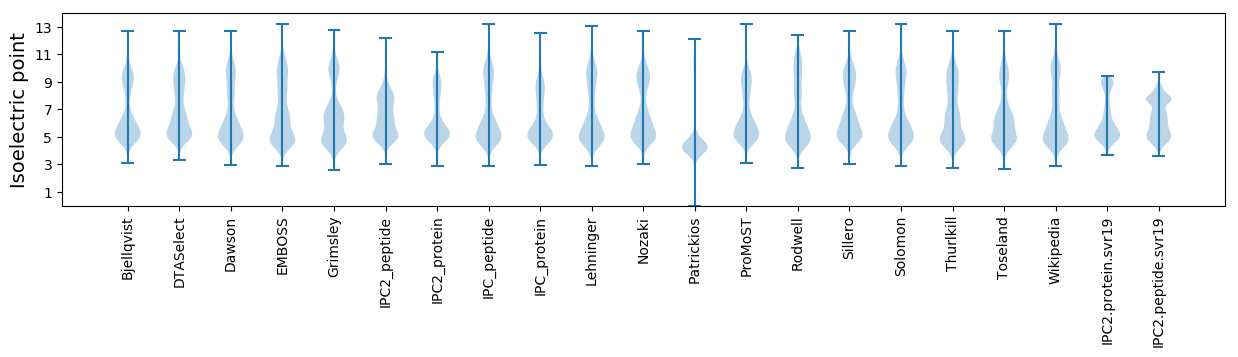
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N6N5U1|A0A6N6N5U1_9DELT Histidine kinase OS=Desulfovibrio senegalensis OX=1721087 GN=F8A88_00810 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.98TRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.99RR14 pKa = 11.84THH16 pKa = 5.86GFLVRR21 pKa = 11.84SRR23 pKa = 11.84TKK25 pKa = 10.5NGRR28 pKa = 11.84KK29 pKa = 9.22VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.8GRR39 pKa = 11.84KK40 pKa = 8.66RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.98TRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.99RR14 pKa = 11.84THH16 pKa = 5.86GFLVRR21 pKa = 11.84SRR23 pKa = 11.84TKK25 pKa = 10.5NGRR28 pKa = 11.84KK29 pKa = 9.22VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.8GRR39 pKa = 11.84KK40 pKa = 8.66RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1003229 |
28 |
2808 |
327.6 |
36.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.529 ± 0.059 | 1.405 ± 0.02 |
5.828 ± 0.037 | 6.503 ± 0.046 |
4.067 ± 0.029 | 7.946 ± 0.045 |
2.21 ± 0.021 | 5.194 ± 0.037 |
4.608 ± 0.045 | 10.084 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.18 ± 0.021 | 3.278 ± 0.025 |
4.584 ± 0.029 | 3.355 ± 0.025 |
6.293 ± 0.049 | 5.549 ± 0.027 |
5.081 ± 0.033 | 7.531 ± 0.038 |
1.171 ± 0.018 | 2.603 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
