
Thalassobium sp. R2A62
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Thalassobium; unclassified Thalassobium
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
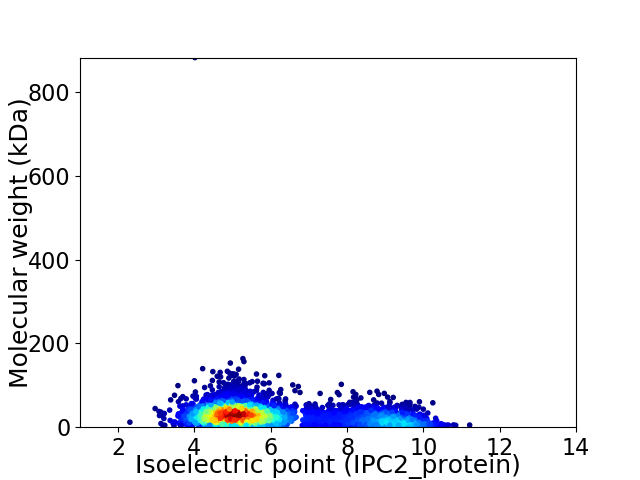
Virtual 2D-PAGE plot for 3672 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
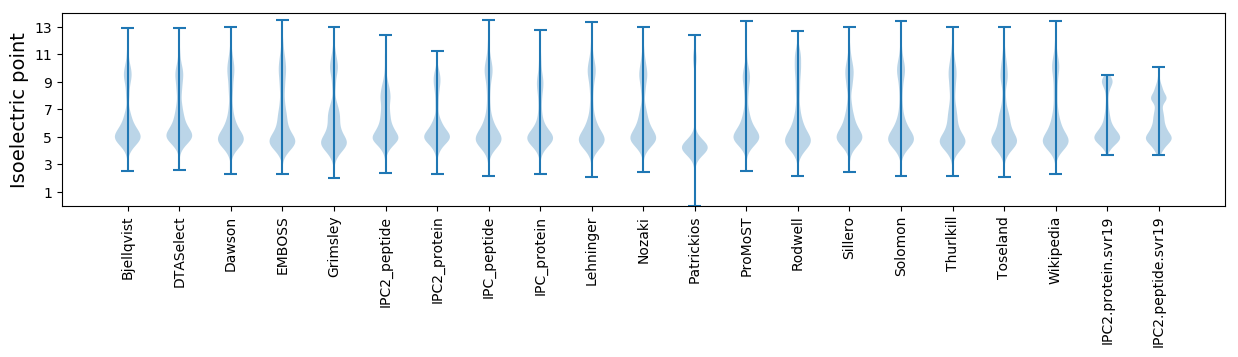
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7DAZ9|C7DAZ9_9RHOB Ankyrin repeat protein OS=Thalassobium sp. R2A62 OX=633131 GN=TR2A62_1737 PE=4 SV=1
MM1 pKa = 7.85AIYY4 pKa = 9.83TFTIYY9 pKa = 10.3QATSITTGPITLSGNAVTITVDD31 pKa = 5.39DD32 pKa = 5.08PDD34 pKa = 4.72DD35 pKa = 3.92SQGIGDD41 pKa = 5.37LLTDD45 pKa = 4.32GSGHH49 pKa = 6.2DD50 pKa = 4.09VEE52 pKa = 5.3TGITPSITAVSDD64 pKa = 3.37PAYY67 pKa = 10.63SYY69 pKa = 11.28LIGDD73 pKa = 4.26GLTFNGQWAIDD84 pKa = 3.96GLSGTGWNALILNDD98 pKa = 4.36EE99 pKa = 4.92DD100 pKa = 4.21GSNNASFHH108 pKa = 4.82GTMWLASHH116 pKa = 7.63PDD118 pKa = 3.6NPTALADD125 pKa = 3.8AFISGSNVGIGSRR138 pKa = 11.84TDD140 pKa = 3.0VDD142 pKa = 4.18SVPEE146 pKa = 4.3LVPPAPVEE154 pKa = 4.06TYY156 pKa = 9.57TFHH159 pKa = 6.84TFSQSDD165 pKa = 3.41VGFPQTSAFSKK176 pKa = 10.26DD177 pKa = 3.38GIVFVHH183 pKa = 7.4DD184 pKa = 3.59STNPITTIAVDD195 pKa = 4.25DD196 pKa = 5.0DD197 pKa = 4.61DD198 pKa = 6.32AVLDD202 pKa = 4.33DD203 pKa = 4.2QSTNDD208 pKa = 3.25GQTYY212 pKa = 10.22DD213 pKa = 3.51SSEE216 pKa = 3.77ILQSVNGDD224 pKa = 3.42TSISGNDD231 pKa = 3.02IASFASYY238 pKa = 9.63TVTGDD243 pKa = 3.51DD244 pKa = 3.54GSSFKK249 pKa = 10.95AYY251 pKa = 9.76IIAGSNGTNIRR262 pKa = 11.84DD263 pKa = 3.48IDD265 pKa = 3.9GSALTGFDD273 pKa = 3.28TYY275 pKa = 10.61ITFTQPLIDD284 pKa = 3.86GVEE287 pKa = 4.17YY288 pKa = 9.49TYY290 pKa = 11.28SEE292 pKa = 4.49YY293 pKa = 11.41SPIGQVAYY301 pKa = 10.62ADD303 pKa = 4.02LAEE306 pKa = 4.61QGTFDD311 pKa = 4.48IVGTPPEE318 pKa = 4.31SPEE321 pKa = 4.33AGLSISEE328 pKa = 4.94AIFLGNFMDD337 pKa = 5.24ADD339 pKa = 3.65TSEE342 pKa = 4.66EE343 pKa = 4.08FFPDD347 pKa = 3.67TVNTAPFLGTFGSSSDD363 pKa = 4.26PLWQDD368 pKa = 3.08EE369 pKa = 4.53VLVTYY374 pKa = 10.75EE375 pKa = 4.54DD376 pKa = 3.95VNGDD380 pKa = 3.53GAINTDD386 pKa = 3.52NLSPQYY392 pKa = 10.96LPDD395 pKa = 3.61EE396 pKa = 4.91EE397 pKa = 4.5ISYY400 pKa = 11.18DD401 pKa = 3.97LGSGTQTALVDD412 pKa = 4.35SIVSINLTVTYY423 pKa = 10.03TDD425 pKa = 4.55GSTQSYY431 pKa = 9.28TNALMYY437 pKa = 10.46QDD439 pKa = 5.03DD440 pKa = 4.58AGNMFLVNSDD450 pKa = 3.09WAGTNLNSGSWPQIQSIDD468 pKa = 3.69VTSVTDD474 pKa = 5.08GYY476 pKa = 10.36HH477 pKa = 6.59GAMTQDD483 pKa = 3.89DD484 pKa = 4.38FQSFVCFTRR493 pKa = 11.84GTLIKK498 pKa = 9.04TDD500 pKa = 3.18QGEE503 pKa = 4.16RR504 pKa = 11.84PIEE507 pKa = 3.89EE508 pKa = 4.16LAAGDD513 pKa = 3.69MVLTMDD519 pKa = 5.11HH520 pKa = 7.25GYY522 pKa = 10.36QPIRR526 pKa = 11.84WIGSSKK532 pKa = 10.35RR533 pKa = 11.84AATGDD538 pKa = 3.71LAPILIRR545 pKa = 11.84RR546 pKa = 11.84GALGNDD552 pKa = 3.11RR553 pKa = 11.84DD554 pKa = 4.18LRR556 pKa = 11.84VSPQHH561 pKa = 7.28RR562 pKa = 11.84MLLQGWQAEE571 pKa = 4.29LLFGEE576 pKa = 4.69VEE578 pKa = 4.33VLATAKK584 pKa = 10.68SLLNDD589 pKa = 3.24QTILRR594 pKa = 11.84DD595 pKa = 3.33EE596 pKa = 4.68GGEE599 pKa = 3.86VEE601 pKa = 4.37YY602 pKa = 10.93FHH604 pKa = 6.82MLFDD608 pKa = 3.52THH610 pKa = 6.63EE611 pKa = 4.42IIYY614 pKa = 10.9AEE616 pKa = 4.53GCPSEE621 pKa = 4.83SFHH624 pKa = 7.24PGQQGWKK631 pKa = 10.1ALDD634 pKa = 3.18QATRR638 pKa = 11.84DD639 pKa = 3.74EE640 pKa = 4.55ILTLFPQLVDD650 pKa = 3.52GTLNDD655 pKa = 3.72YY656 pKa = 11.0GPAARR661 pKa = 11.84MSLKK665 pKa = 10.48HH666 pKa = 6.62KK667 pKa = 9.91EE668 pKa = 4.05GKK670 pKa = 10.38LLGSYY675 pKa = 8.52MAGSVV680 pKa = 3.13
MM1 pKa = 7.85AIYY4 pKa = 9.83TFTIYY9 pKa = 10.3QATSITTGPITLSGNAVTITVDD31 pKa = 5.39DD32 pKa = 5.08PDD34 pKa = 4.72DD35 pKa = 3.92SQGIGDD41 pKa = 5.37LLTDD45 pKa = 4.32GSGHH49 pKa = 6.2DD50 pKa = 4.09VEE52 pKa = 5.3TGITPSITAVSDD64 pKa = 3.37PAYY67 pKa = 10.63SYY69 pKa = 11.28LIGDD73 pKa = 4.26GLTFNGQWAIDD84 pKa = 3.96GLSGTGWNALILNDD98 pKa = 4.36EE99 pKa = 4.92DD100 pKa = 4.21GSNNASFHH108 pKa = 4.82GTMWLASHH116 pKa = 7.63PDD118 pKa = 3.6NPTALADD125 pKa = 3.8AFISGSNVGIGSRR138 pKa = 11.84TDD140 pKa = 3.0VDD142 pKa = 4.18SVPEE146 pKa = 4.3LVPPAPVEE154 pKa = 4.06TYY156 pKa = 9.57TFHH159 pKa = 6.84TFSQSDD165 pKa = 3.41VGFPQTSAFSKK176 pKa = 10.26DD177 pKa = 3.38GIVFVHH183 pKa = 7.4DD184 pKa = 3.59STNPITTIAVDD195 pKa = 4.25DD196 pKa = 5.0DD197 pKa = 4.61DD198 pKa = 6.32AVLDD202 pKa = 4.33DD203 pKa = 4.2QSTNDD208 pKa = 3.25GQTYY212 pKa = 10.22DD213 pKa = 3.51SSEE216 pKa = 3.77ILQSVNGDD224 pKa = 3.42TSISGNDD231 pKa = 3.02IASFASYY238 pKa = 9.63TVTGDD243 pKa = 3.51DD244 pKa = 3.54GSSFKK249 pKa = 10.95AYY251 pKa = 9.76IIAGSNGTNIRR262 pKa = 11.84DD263 pKa = 3.48IDD265 pKa = 3.9GSALTGFDD273 pKa = 3.28TYY275 pKa = 10.61ITFTQPLIDD284 pKa = 3.86GVEE287 pKa = 4.17YY288 pKa = 9.49TYY290 pKa = 11.28SEE292 pKa = 4.49YY293 pKa = 11.41SPIGQVAYY301 pKa = 10.62ADD303 pKa = 4.02LAEE306 pKa = 4.61QGTFDD311 pKa = 4.48IVGTPPEE318 pKa = 4.31SPEE321 pKa = 4.33AGLSISEE328 pKa = 4.94AIFLGNFMDD337 pKa = 5.24ADD339 pKa = 3.65TSEE342 pKa = 4.66EE343 pKa = 4.08FFPDD347 pKa = 3.67TVNTAPFLGTFGSSSDD363 pKa = 4.26PLWQDD368 pKa = 3.08EE369 pKa = 4.53VLVTYY374 pKa = 10.75EE375 pKa = 4.54DD376 pKa = 3.95VNGDD380 pKa = 3.53GAINTDD386 pKa = 3.52NLSPQYY392 pKa = 10.96LPDD395 pKa = 3.61EE396 pKa = 4.91EE397 pKa = 4.5ISYY400 pKa = 11.18DD401 pKa = 3.97LGSGTQTALVDD412 pKa = 4.35SIVSINLTVTYY423 pKa = 10.03TDD425 pKa = 4.55GSTQSYY431 pKa = 9.28TNALMYY437 pKa = 10.46QDD439 pKa = 5.03DD440 pKa = 4.58AGNMFLVNSDD450 pKa = 3.09WAGTNLNSGSWPQIQSIDD468 pKa = 3.69VTSVTDD474 pKa = 5.08GYY476 pKa = 10.36HH477 pKa = 6.59GAMTQDD483 pKa = 3.89DD484 pKa = 4.38FQSFVCFTRR493 pKa = 11.84GTLIKK498 pKa = 9.04TDD500 pKa = 3.18QGEE503 pKa = 4.16RR504 pKa = 11.84PIEE507 pKa = 3.89EE508 pKa = 4.16LAAGDD513 pKa = 3.69MVLTMDD519 pKa = 5.11HH520 pKa = 7.25GYY522 pKa = 10.36QPIRR526 pKa = 11.84WIGSSKK532 pKa = 10.35RR533 pKa = 11.84AATGDD538 pKa = 3.71LAPILIRR545 pKa = 11.84RR546 pKa = 11.84GALGNDD552 pKa = 3.11RR553 pKa = 11.84DD554 pKa = 4.18LRR556 pKa = 11.84VSPQHH561 pKa = 7.28RR562 pKa = 11.84MLLQGWQAEE571 pKa = 4.29LLFGEE576 pKa = 4.69VEE578 pKa = 4.33VLATAKK584 pKa = 10.68SLLNDD589 pKa = 3.24QTILRR594 pKa = 11.84DD595 pKa = 3.33EE596 pKa = 4.68GGEE599 pKa = 3.86VEE601 pKa = 4.37YY602 pKa = 10.93FHH604 pKa = 6.82MLFDD608 pKa = 3.52THH610 pKa = 6.63EE611 pKa = 4.42IIYY614 pKa = 10.9AEE616 pKa = 4.53GCPSEE621 pKa = 4.83SFHH624 pKa = 7.24PGQQGWKK631 pKa = 10.1ALDD634 pKa = 3.18QATRR638 pKa = 11.84DD639 pKa = 3.74EE640 pKa = 4.55ILTLFPQLVDD650 pKa = 3.52GTLNDD655 pKa = 3.72YY656 pKa = 11.0GPAARR661 pKa = 11.84MSLKK665 pKa = 10.48HH666 pKa = 6.62KK667 pKa = 9.91EE668 pKa = 4.05GKK670 pKa = 10.38LLGSYY675 pKa = 8.52MAGSVV680 pKa = 3.13
Molecular weight: 72.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7D7M6|C7D7M6_9RHOB Phage integrase OS=Thalassobium sp. R2A62 OX=633131 GN=TR2A62_3212 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1040251 |
33 |
8645 |
283.3 |
30.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.412 ± 0.051 | 0.937 ± 0.015 |
6.477 ± 0.038 | 5.621 ± 0.045 |
3.904 ± 0.03 | 8.408 ± 0.058 |
2.071 ± 0.025 | 5.595 ± 0.031 |
3.45 ± 0.037 | 9.535 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.969 ± 0.027 | 3.061 ± 0.03 |
4.643 ± 0.037 | 3.357 ± 0.024 |
6.059 ± 0.051 | 5.498 ± 0.038 |
5.944 ± 0.042 | 7.345 ± 0.039 |
1.389 ± 0.019 | 2.322 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
