
Cryobacterium psychrotolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Cryobacterium
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
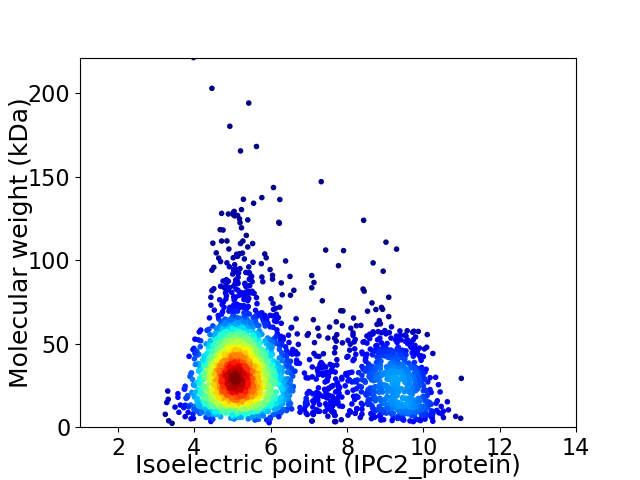
Virtual 2D-PAGE plot for 3047 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G8ZC01|A0A1G8ZC01_9MICO Carboxypeptidase regulatory-like domain-containing protein OS=Cryobacterium psychrotolerans OX=386301 GN=E3T56_06240 PE=4 SV=1
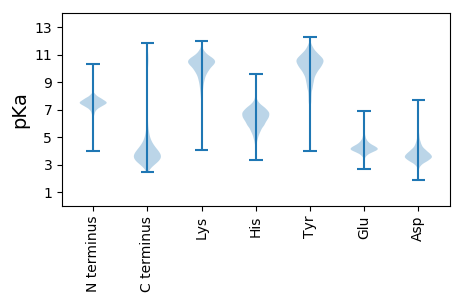
MM1 pKa = 7.12NRR3 pKa = 11.84SASKK7 pKa = 8.84FTMVGLMVSGILFAGTACASNQSGADD33 pKa = 3.49SAAGPAGDD41 pKa = 4.23VAPSSSPGPEE51 pKa = 3.59VDD53 pKa = 3.75PLTSVTTIVVRR64 pKa = 11.84PEE66 pKa = 3.44ALEE69 pKa = 4.55LMDD72 pKa = 4.34EE73 pKa = 4.46SGSLVASLTYY83 pKa = 10.25EE84 pKa = 4.39DD85 pKa = 5.16PLAAMLGALSVVVGEE100 pKa = 4.75EE101 pKa = 3.93PATASYY107 pKa = 10.88EE108 pKa = 4.31GGLEE112 pKa = 3.96RR113 pKa = 11.84PAGQTYY119 pKa = 9.73SWAGLTLTDD128 pKa = 4.01YY129 pKa = 11.04LPPEE133 pKa = 4.39GKK135 pKa = 10.26FLGEE139 pKa = 3.87SDD141 pKa = 3.91YY142 pKa = 11.85SLDD145 pKa = 3.49FTAGQVGDD153 pKa = 3.97GVAITTEE160 pKa = 4.15SGSTVGDD167 pKa = 3.86DD168 pKa = 3.31MDD170 pKa = 4.6AVADD174 pKa = 4.03EE175 pKa = 5.33LGLSVNRR182 pKa = 11.84DD183 pKa = 3.41FASAGHH189 pKa = 7.0LFFLIEE195 pKa = 4.24VGPEE199 pKa = 3.71LTTPAGDD206 pKa = 3.57NIEE209 pKa = 4.58YY210 pKa = 8.98PNAWAVHH217 pKa = 5.08VSSTSGSGVINNIGAPVNLSHH238 pKa = 6.8WVSS241 pKa = 3.29
MM1 pKa = 7.12NRR3 pKa = 11.84SASKK7 pKa = 8.84FTMVGLMVSGILFAGTACASNQSGADD33 pKa = 3.49SAAGPAGDD41 pKa = 4.23VAPSSSPGPEE51 pKa = 3.59VDD53 pKa = 3.75PLTSVTTIVVRR64 pKa = 11.84PEE66 pKa = 3.44ALEE69 pKa = 4.55LMDD72 pKa = 4.34EE73 pKa = 4.46SGSLVASLTYY83 pKa = 10.25EE84 pKa = 4.39DD85 pKa = 5.16PLAAMLGALSVVVGEE100 pKa = 4.75EE101 pKa = 3.93PATASYY107 pKa = 10.88EE108 pKa = 4.31GGLEE112 pKa = 3.96RR113 pKa = 11.84PAGQTYY119 pKa = 9.73SWAGLTLTDD128 pKa = 4.01YY129 pKa = 11.04LPPEE133 pKa = 4.39GKK135 pKa = 10.26FLGEE139 pKa = 3.87SDD141 pKa = 3.91YY142 pKa = 11.85SLDD145 pKa = 3.49FTAGQVGDD153 pKa = 3.97GVAITTEE160 pKa = 4.15SGSTVGDD167 pKa = 3.86DD168 pKa = 3.31MDD170 pKa = 4.6AVADD174 pKa = 4.03EE175 pKa = 5.33LGLSVNRR182 pKa = 11.84DD183 pKa = 3.41FASAGHH189 pKa = 7.0LFFLIEE195 pKa = 4.24VGPEE199 pKa = 3.71LTTPAGDD206 pKa = 3.57NIEE209 pKa = 4.58YY210 pKa = 8.98PNAWAVHH217 pKa = 5.08VSSTSGSGVINNIGAPVNLSHH238 pKa = 6.8WVSS241 pKa = 3.29
Molecular weight: 24.52 kDa
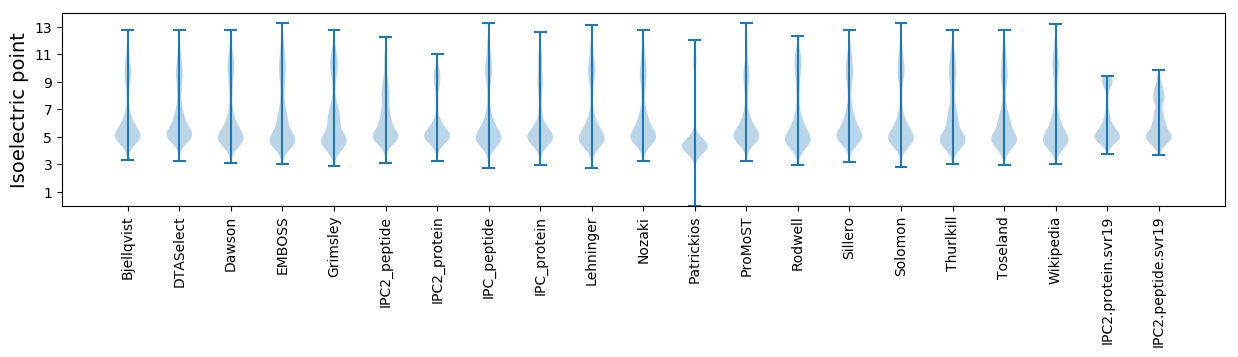
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9FNI6|A0A1G9FNI6_9MICO Uncharacterized protein OS=Cryobacterium psychrotolerans OX=386301 GN=SAMN05216282_11727 PE=4 SV=1
MM1 pKa = 7.45LRR3 pKa = 11.84TAATQTVPPSRR14 pKa = 11.84VTVSVRR20 pKa = 11.84LIAGTPTVHH29 pKa = 7.21SVMASVLLIAATPTGPRR46 pKa = 11.84NRR48 pKa = 11.84ATVNASPGRR57 pKa = 11.84RR58 pKa = 11.84MGSVRR63 pKa = 11.84LTAEE67 pKa = 4.0TPIVRR72 pKa = 11.84SVMASVLLTVATPTVPPSPVSASRR96 pKa = 11.84GRR98 pKa = 11.84RR99 pKa = 11.84MATARR104 pKa = 11.84LTAEE108 pKa = 4.17TPTVRR113 pKa = 11.84SRR115 pKa = 11.84AATAPPTGATRR126 pKa = 11.84TVPRR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84VMVSASRR139 pKa = 11.84GRR141 pKa = 11.84RR142 pKa = 11.84MASGLRR148 pKa = 11.84SATLTVRR155 pKa = 11.84PIAPMPRR162 pKa = 11.84VEE164 pKa = 4.4SVRR167 pKa = 11.84SVMVSASPGPRR178 pKa = 11.84MASVPRR184 pKa = 11.84IVATPIVRR192 pKa = 11.84SRR194 pKa = 11.84VASVPRR200 pKa = 11.84TGEE203 pKa = 3.87TRR205 pKa = 11.84TVPPSRR211 pKa = 11.84VTASASPGPGMVPRR225 pKa = 11.84PSARR229 pKa = 11.84AVTLLPVTGNARR241 pKa = 11.84SGAHH245 pKa = 5.25AAVRR249 pKa = 11.84RR250 pKa = 11.84PVPAPPVTPEE260 pKa = 4.17KK261 pKa = 10.92SSGPAKK267 pKa = 10.36APPPAVTATRR277 pKa = 11.84APSKK281 pKa = 10.57RR282 pKa = 11.84RR283 pKa = 3.41
MM1 pKa = 7.45LRR3 pKa = 11.84TAATQTVPPSRR14 pKa = 11.84VTVSVRR20 pKa = 11.84LIAGTPTVHH29 pKa = 7.21SVMASVLLIAATPTGPRR46 pKa = 11.84NRR48 pKa = 11.84ATVNASPGRR57 pKa = 11.84RR58 pKa = 11.84MGSVRR63 pKa = 11.84LTAEE67 pKa = 4.0TPIVRR72 pKa = 11.84SVMASVLLTVATPTVPPSPVSASRR96 pKa = 11.84GRR98 pKa = 11.84RR99 pKa = 11.84MATARR104 pKa = 11.84LTAEE108 pKa = 4.17TPTVRR113 pKa = 11.84SRR115 pKa = 11.84AATAPPTGATRR126 pKa = 11.84TVPRR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84VMVSASRR139 pKa = 11.84GRR141 pKa = 11.84RR142 pKa = 11.84MASGLRR148 pKa = 11.84SATLTVRR155 pKa = 11.84PIAPMPRR162 pKa = 11.84VEE164 pKa = 4.4SVRR167 pKa = 11.84SVMVSASPGPRR178 pKa = 11.84MASVPRR184 pKa = 11.84IVATPIVRR192 pKa = 11.84SRR194 pKa = 11.84VASVPRR200 pKa = 11.84TGEE203 pKa = 3.87TRR205 pKa = 11.84TVPPSRR211 pKa = 11.84VTASASPGPGMVPRR225 pKa = 11.84PSARR229 pKa = 11.84AVTLLPVTGNARR241 pKa = 11.84SGAHH245 pKa = 5.25AAVRR249 pKa = 11.84RR250 pKa = 11.84PVPAPPVTPEE260 pKa = 4.17KK261 pKa = 10.92SSGPAKK267 pKa = 10.36APPPAVTATRR277 pKa = 11.84APSKK281 pKa = 10.57RR282 pKa = 11.84RR283 pKa = 3.41
Molecular weight: 29.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
978152 |
25 |
2218 |
321.0 |
34.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.252 ± 0.064 | 0.533 ± 0.011 |
5.842 ± 0.038 | 5.457 ± 0.04 |
3.179 ± 0.025 | 9.053 ± 0.039 |
1.985 ± 0.022 | 4.723 ± 0.032 |
2.287 ± 0.03 | 10.438 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.845 ± 0.017 | 2.219 ± 0.026 |
5.382 ± 0.029 | 2.802 ± 0.021 |
6.986 ± 0.044 | 5.856 ± 0.03 |
6.075 ± 0.046 | 8.645 ± 0.042 |
1.413 ± 0.018 | 2.025 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
