
Capybara microvirus Cap1_SP_127
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
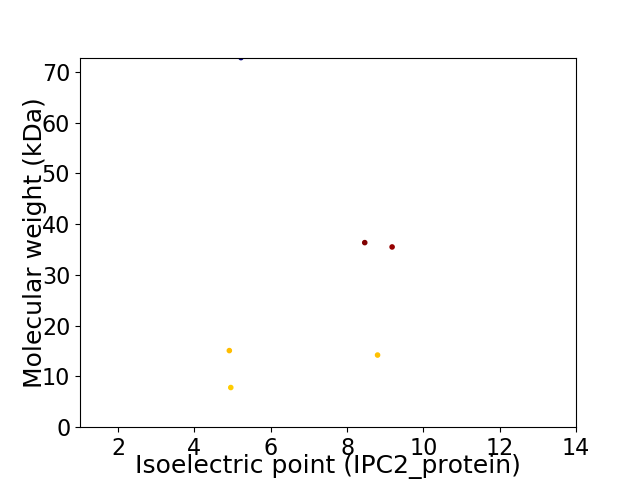
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5B3|A0A4P8W5B3_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_127 OX=2585380 PE=4 SV=1
MM1 pKa = 7.79TYY3 pKa = 11.04NDD5 pKa = 3.94YY6 pKa = 10.74LQQYY10 pKa = 9.22AHH12 pKa = 7.48LFHH15 pKa = 7.08EE16 pKa = 5.45KK17 pKa = 9.86YY18 pKa = 10.7DD19 pKa = 3.77KK20 pKa = 11.03VRR22 pKa = 11.84NTIPDD27 pKa = 3.85AVPSDD32 pKa = 4.0SQLVAQFTGKK42 pKa = 10.06VPLTSNTGQIGVQKK56 pKa = 10.42EE57 pKa = 4.81SYY59 pKa = 9.45QSTYY63 pKa = 11.01QEE65 pKa = 4.12VNLDD69 pKa = 3.91NILPEE74 pKa = 4.37SKK76 pKa = 10.59LYY78 pKa = 8.06MTFEE82 pKa = 4.78DD83 pKa = 4.04AFQNLRR89 pKa = 11.84DD90 pKa = 4.07GGNAATNLRR99 pKa = 11.84DD100 pKa = 3.36LNEE103 pKa = 4.61YY104 pKa = 7.93YY105 pKa = 9.63TQLKK109 pKa = 10.19NKK111 pKa = 9.59EE112 pKa = 3.91KK113 pKa = 10.69SQNLNPADD121 pKa = 3.95GSTATSSNQDD131 pKa = 2.8NKK133 pKa = 11.39
MM1 pKa = 7.79TYY3 pKa = 11.04NDD5 pKa = 3.94YY6 pKa = 10.74LQQYY10 pKa = 9.22AHH12 pKa = 7.48LFHH15 pKa = 7.08EE16 pKa = 5.45KK17 pKa = 9.86YY18 pKa = 10.7DD19 pKa = 3.77KK20 pKa = 11.03VRR22 pKa = 11.84NTIPDD27 pKa = 3.85AVPSDD32 pKa = 4.0SQLVAQFTGKK42 pKa = 10.06VPLTSNTGQIGVQKK56 pKa = 10.42EE57 pKa = 4.81SYY59 pKa = 9.45QSTYY63 pKa = 11.01QEE65 pKa = 4.12VNLDD69 pKa = 3.91NILPEE74 pKa = 4.37SKK76 pKa = 10.59LYY78 pKa = 8.06MTFEE82 pKa = 4.78DD83 pKa = 4.04AFQNLRR89 pKa = 11.84DD90 pKa = 4.07GGNAATNLRR99 pKa = 11.84DD100 pKa = 3.36LNEE103 pKa = 4.61YY104 pKa = 7.93YY105 pKa = 9.63TQLKK109 pKa = 10.19NKK111 pKa = 9.59EE112 pKa = 3.91KK113 pKa = 10.69SQNLNPADD121 pKa = 3.95GSTATSSNQDD131 pKa = 2.8NKK133 pKa = 11.39
Molecular weight: 15.1 kDa
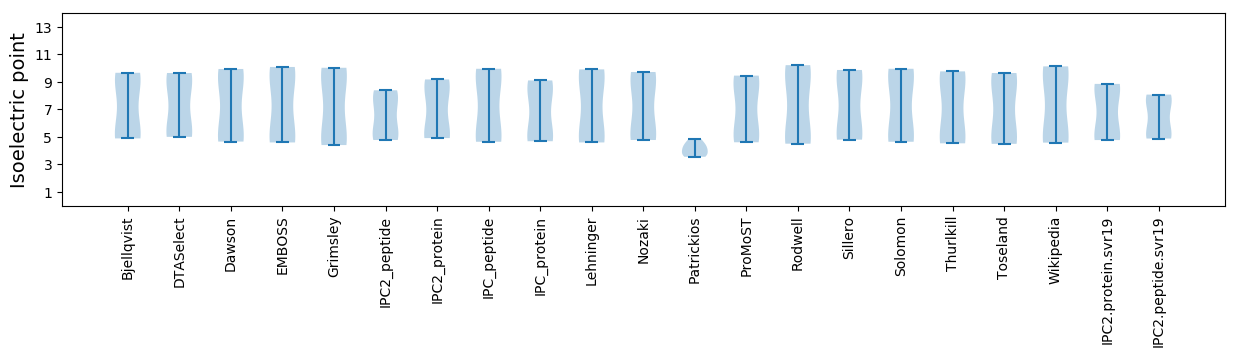
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W474|A0A4P8W474_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_127 OX=2585380 PE=4 SV=1
MM1 pKa = 7.27NCLHH5 pKa = 6.6PVDD8 pKa = 4.53VRR10 pKa = 11.84ITRR13 pKa = 11.84KK14 pKa = 10.44DD15 pKa = 3.12GTLIPALVPCGKK27 pKa = 10.39CYY29 pKa = 10.58ACQYY33 pKa = 8.98NDD35 pKa = 3.38RR36 pKa = 11.84VGWSYY41 pKa = 11.35RR42 pKa = 11.84ATLEE46 pKa = 4.43SNSSKK51 pKa = 10.86YY52 pKa = 10.91SLFVTLTYY60 pKa = 11.46NNLHH64 pKa = 6.66LDD66 pKa = 3.64KK67 pKa = 10.01LTHH70 pKa = 6.14NGKK73 pKa = 10.3SYY75 pKa = 10.91GVVSRR80 pKa = 11.84EE81 pKa = 3.63TLQKK85 pKa = 10.36FFKK88 pKa = 10.08RR89 pKa = 11.84CRR91 pKa = 11.84KK92 pKa = 9.69RR93 pKa = 11.84GLKK96 pKa = 9.98FSYY99 pKa = 10.19IFVSEE104 pKa = 4.08YY105 pKa = 11.19GPLTNRR111 pKa = 11.84PHH113 pKa = 4.98YY114 pKa = 10.36HH115 pKa = 6.8GIFFLHH121 pKa = 6.4QDD123 pKa = 3.19NTFEE127 pKa = 5.5LDD129 pKa = 4.43DD130 pKa = 5.0IIEE133 pKa = 4.87KK134 pKa = 10.24ISKK137 pKa = 9.16SWNLGFSKK145 pKa = 10.16ISEE148 pKa = 4.22VTPARR153 pKa = 11.84INYY156 pKa = 5.23VTGYY160 pKa = 9.76SFKK163 pKa = 10.47EE164 pKa = 4.15CNPKK168 pKa = 10.25FSFQTPTFRR177 pKa = 11.84LMSRR181 pKa = 11.84RR182 pKa = 11.84PAIGYY187 pKa = 9.01DD188 pKa = 3.69LLADD192 pKa = 3.78EE193 pKa = 4.31QFKK196 pKa = 11.15NISNHH201 pKa = 5.31IANGGNSLRR210 pKa = 11.84FGSASIPIPKK220 pKa = 9.75YY221 pKa = 10.42YY222 pKa = 10.31KK223 pKa = 9.08NKK225 pKa = 9.71LRR227 pKa = 11.84KK228 pKa = 8.86EE229 pKa = 4.11HH230 pKa = 6.52TNDD233 pKa = 3.08EE234 pKa = 4.38LFNIYY239 pKa = 10.27NSQITRR245 pKa = 11.84QNLRR249 pKa = 11.84TKK251 pKa = 10.64KK252 pKa = 10.07LFEE255 pKa = 4.27YY256 pKa = 9.98FQSKK260 pKa = 9.44YY261 pKa = 10.22PKK263 pKa = 9.84LSPSRR268 pKa = 11.84IYY270 pKa = 11.11SLIYY274 pKa = 10.51SDD276 pKa = 5.92DD277 pKa = 3.71DD278 pKa = 3.7TNLRR282 pKa = 11.84LKK284 pKa = 10.6RR285 pKa = 11.84QALKK289 pKa = 10.86NIYY292 pKa = 10.14KK293 pKa = 10.08NLKK296 pKa = 9.39NNEE299 pKa = 4.16DD300 pKa = 3.63NFNN303 pKa = 3.58
MM1 pKa = 7.27NCLHH5 pKa = 6.6PVDD8 pKa = 4.53VRR10 pKa = 11.84ITRR13 pKa = 11.84KK14 pKa = 10.44DD15 pKa = 3.12GTLIPALVPCGKK27 pKa = 10.39CYY29 pKa = 10.58ACQYY33 pKa = 8.98NDD35 pKa = 3.38RR36 pKa = 11.84VGWSYY41 pKa = 11.35RR42 pKa = 11.84ATLEE46 pKa = 4.43SNSSKK51 pKa = 10.86YY52 pKa = 10.91SLFVTLTYY60 pKa = 11.46NNLHH64 pKa = 6.66LDD66 pKa = 3.64KK67 pKa = 10.01LTHH70 pKa = 6.14NGKK73 pKa = 10.3SYY75 pKa = 10.91GVVSRR80 pKa = 11.84EE81 pKa = 3.63TLQKK85 pKa = 10.36FFKK88 pKa = 10.08RR89 pKa = 11.84CRR91 pKa = 11.84KK92 pKa = 9.69RR93 pKa = 11.84GLKK96 pKa = 9.98FSYY99 pKa = 10.19IFVSEE104 pKa = 4.08YY105 pKa = 11.19GPLTNRR111 pKa = 11.84PHH113 pKa = 4.98YY114 pKa = 10.36HH115 pKa = 6.8GIFFLHH121 pKa = 6.4QDD123 pKa = 3.19NTFEE127 pKa = 5.5LDD129 pKa = 4.43DD130 pKa = 5.0IIEE133 pKa = 4.87KK134 pKa = 10.24ISKK137 pKa = 9.16SWNLGFSKK145 pKa = 10.16ISEE148 pKa = 4.22VTPARR153 pKa = 11.84INYY156 pKa = 5.23VTGYY160 pKa = 9.76SFKK163 pKa = 10.47EE164 pKa = 4.15CNPKK168 pKa = 10.25FSFQTPTFRR177 pKa = 11.84LMSRR181 pKa = 11.84RR182 pKa = 11.84PAIGYY187 pKa = 9.01DD188 pKa = 3.69LLADD192 pKa = 3.78EE193 pKa = 4.31QFKK196 pKa = 11.15NISNHH201 pKa = 5.31IANGGNSLRR210 pKa = 11.84FGSASIPIPKK220 pKa = 9.75YY221 pKa = 10.42YY222 pKa = 10.31KK223 pKa = 9.08NKK225 pKa = 9.71LRR227 pKa = 11.84KK228 pKa = 8.86EE229 pKa = 4.11HH230 pKa = 6.52TNDD233 pKa = 3.08EE234 pKa = 4.38LFNIYY239 pKa = 10.27NSQITRR245 pKa = 11.84QNLRR249 pKa = 11.84TKK251 pKa = 10.64KK252 pKa = 10.07LFEE255 pKa = 4.27YY256 pKa = 9.98FQSKK260 pKa = 9.44YY261 pKa = 10.22PKK263 pKa = 9.84LSPSRR268 pKa = 11.84IYY270 pKa = 11.11SLIYY274 pKa = 10.51SDD276 pKa = 5.92DD277 pKa = 3.71DD278 pKa = 3.7TNLRR282 pKa = 11.84LKK284 pKa = 10.6RR285 pKa = 11.84QALKK289 pKa = 10.86NIYY292 pKa = 10.14KK293 pKa = 10.08NLKK296 pKa = 9.39NNEE299 pKa = 4.16DD300 pKa = 3.63NFNN303 pKa = 3.58
Molecular weight: 35.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1575 |
69 |
636 |
262.5 |
30.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.254 ± 0.594 | 1.143 ± 0.595 |
5.587 ± 0.577 | 5.016 ± 0.313 |
6.095 ± 1.03 | 4.889 ± 0.429 |
1.651 ± 0.32 | 5.841 ± 0.906 |
7.111 ± 1.691 | 9.143 ± 0.61 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.587 ± 0.329 | 8.571 ± 0.989 |
4.635 ± 0.709 | 5.016 ± 1.167 |
4.508 ± 0.775 | 7.937 ± 0.394 |
6.222 ± 0.775 | 4.127 ± 0.732 |
0.762 ± 0.153 | 5.905 ± 0.925 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
