
Sclerotinia sclerotiorum mitovirus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.85
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
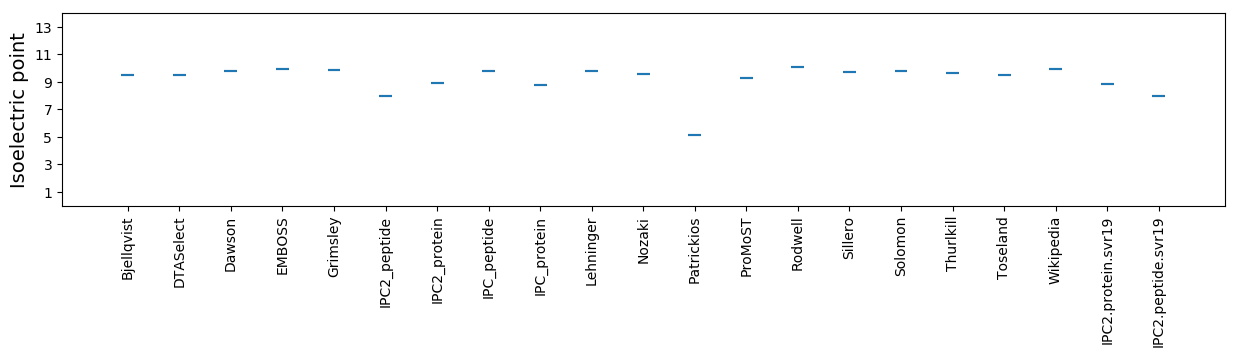
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0S4GCV3|A0A0S4GCV3_9VIRU RNA dependent RNA polymerase OS=Sclerotinia sclerotiorum mitovirus 3 OX=1279099 GN=RdRp PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.03HH3 pKa = 6.45LKK5 pKa = 8.74LTTYY9 pKa = 10.56KK10 pKa = 10.76HH11 pKa = 6.66LDD13 pKa = 3.53LNINQADD20 pKa = 4.12RR21 pKa = 11.84FSNGAFAALSLKK33 pKa = 10.13LWYY36 pKa = 9.22PYY38 pKa = 10.55VRR40 pKa = 11.84LLIWSLRR47 pKa = 11.84LDD49 pKa = 3.78KK50 pKa = 10.92PSFMKK55 pKa = 10.34FATRR59 pKa = 11.84IEE61 pKa = 4.33SLWIHH66 pKa = 6.41NGLIFTVKK74 pKa = 10.19YY75 pKa = 10.55LKK77 pKa = 10.31EE78 pKa = 3.9CLRR81 pKa = 11.84ICQHH85 pKa = 5.93FVSGNPVLVTTEE97 pKa = 3.94MPISIVRR104 pKa = 11.84GIPTIIPGSIRR115 pKa = 11.84LQMHH119 pKa = 5.9SRR121 pKa = 11.84DD122 pKa = 3.39LSVIRR127 pKa = 11.84GVLSLLSVFRR137 pKa = 11.84IMKK140 pKa = 9.95IPSTLKK146 pKa = 10.56LEE148 pKa = 4.94SITGPFTGLDD158 pKa = 3.39STLPKK163 pKa = 10.64YY164 pKa = 10.43EE165 pKa = 4.63LIKK168 pKa = 10.38IKK170 pKa = 10.37QVLPVFPQIKK180 pKa = 9.66PIRR183 pKa = 11.84LKK185 pKa = 10.71LLRR188 pKa = 11.84SAGPNCKK195 pKa = 10.08VSMLGIWLDD204 pKa = 3.35IKK206 pKa = 10.92AWSQHH211 pKa = 5.63SNFKK215 pKa = 9.68TLIEE219 pKa = 4.79FIQLFPNYY227 pKa = 10.25VEE229 pKa = 4.21FRR231 pKa = 11.84DD232 pKa = 3.74MLLQEE237 pKa = 4.94AYY239 pKa = 10.35NLRR242 pKa = 11.84LSVPSKK248 pKa = 10.52DD249 pKa = 3.3LHH251 pKa = 6.75LGKK254 pKa = 10.37LAIKK258 pKa = 10.28EE259 pKa = 3.98EE260 pKa = 4.33AAGKK264 pKa = 10.41ARR266 pKa = 11.84VFAITDD272 pKa = 3.99SITQSVMGPISDD284 pKa = 5.79AIFKK288 pKa = 8.81MLRR291 pKa = 11.84QIPMDD296 pKa = 3.32GTFNQSAPLDD306 pKa = 3.66RR307 pKa = 11.84LVQLSKK313 pKa = 11.16DD314 pKa = 3.4GAIAEE319 pKa = 4.42KK320 pKa = 10.9DD321 pKa = 3.71RR322 pKa = 11.84IFYY325 pKa = 10.75SYY327 pKa = 10.86DD328 pKa = 2.84LSAATDD334 pKa = 3.56RR335 pKa = 11.84LPINLQKK342 pKa = 10.85DD343 pKa = 3.3ILSIYY348 pKa = 9.5CGEE351 pKa = 4.22SFASKK356 pKa = 9.95WSILMTDD363 pKa = 3.87RR364 pKa = 11.84DD365 pKa = 3.63WYY367 pKa = 8.87LTKK370 pKa = 10.51EE371 pKa = 4.09KK372 pKa = 10.8RR373 pKa = 11.84NLRR376 pKa = 11.84YY377 pKa = 9.81SVGQPMGALSSWAMLALTHH396 pKa = 6.16HH397 pKa = 7.22FIVGMAANRR406 pKa = 11.84VGKK409 pKa = 10.47LGFNHH414 pKa = 6.26YY415 pKa = 10.85ALLGDD420 pKa = 5.56DD421 pKa = 4.49IVIADD426 pKa = 3.96KK427 pKa = 11.17SVADD431 pKa = 4.06SYY433 pKa = 11.42YY434 pKa = 10.37MIMTEE439 pKa = 3.8ILGVQINLSKK449 pKa = 11.1SLVSTNSFEE458 pKa = 4.1FAKK461 pKa = 10.7RR462 pKa = 11.84LVTLEE467 pKa = 4.31GEE469 pKa = 4.46VTPAGPANILLGLRR483 pKa = 11.84SLNGIPSIILDD494 pKa = 3.88MVNKK498 pKa = 9.33GVPVSEE504 pKa = 4.72EE505 pKa = 4.09SLEE508 pKa = 4.18SWMSTIPTVRR518 pKa = 11.84KK519 pKa = 9.46SQLEE523 pKa = 4.03KK524 pKa = 10.91VKK526 pKa = 10.18WVVKK530 pKa = 10.5GPFGFVPTAEE540 pKa = 4.32GLASFLTMSSSLTPVRR556 pKa = 11.84ANQIIHH562 pKa = 6.58AVRR565 pKa = 11.84YY566 pKa = 7.59TKK568 pKa = 10.03FQWDD572 pKa = 3.26LDD574 pKa = 3.55MWQKK578 pKa = 10.23VVKK581 pKa = 10.7DD582 pKa = 3.89NINYY586 pKa = 9.68RR587 pKa = 11.84ISIEE591 pKa = 3.88EE592 pKa = 4.35LYY594 pKa = 10.89FPVGFQDD601 pKa = 3.46YY602 pKa = 9.58PLDD605 pKa = 3.9FRR607 pKa = 11.84FSPIKK612 pKa = 10.1MEE614 pKa = 4.71IIKK617 pKa = 10.62SLSQDD622 pKa = 3.17LRR624 pKa = 11.84DD625 pKa = 4.42LGGQRR630 pKa = 11.84PTFRR634 pKa = 11.84LIFEE638 pKa = 4.73GPIIMFNYY646 pKa = 8.56YY647 pKa = 9.0RR648 pKa = 11.84QGYY651 pKa = 9.16ASEE654 pKa = 3.7IVNYY658 pKa = 9.88IKK660 pKa = 10.92YY661 pKa = 10.01LVHH664 pKa = 6.42QEE666 pKa = 4.07PEE668 pKa = 4.21FKK670 pKa = 10.31IGGSDD675 pKa = 3.24PFLPRR680 pKa = 11.84NFEE683 pKa = 4.05AFAFSSKK690 pKa = 10.5FKK692 pKa = 9.81GQKK695 pKa = 9.46FFEE698 pKa = 4.37QVRR701 pKa = 11.84EE702 pKa = 4.0HH703 pKa = 6.69LRR705 pKa = 11.84ANQLPMM711 pKa = 5.35
MM1 pKa = 7.42KK2 pKa = 10.03HH3 pKa = 6.45LKK5 pKa = 8.74LTTYY9 pKa = 10.56KK10 pKa = 10.76HH11 pKa = 6.66LDD13 pKa = 3.53LNINQADD20 pKa = 4.12RR21 pKa = 11.84FSNGAFAALSLKK33 pKa = 10.13LWYY36 pKa = 9.22PYY38 pKa = 10.55VRR40 pKa = 11.84LLIWSLRR47 pKa = 11.84LDD49 pKa = 3.78KK50 pKa = 10.92PSFMKK55 pKa = 10.34FATRR59 pKa = 11.84IEE61 pKa = 4.33SLWIHH66 pKa = 6.41NGLIFTVKK74 pKa = 10.19YY75 pKa = 10.55LKK77 pKa = 10.31EE78 pKa = 3.9CLRR81 pKa = 11.84ICQHH85 pKa = 5.93FVSGNPVLVTTEE97 pKa = 3.94MPISIVRR104 pKa = 11.84GIPTIIPGSIRR115 pKa = 11.84LQMHH119 pKa = 5.9SRR121 pKa = 11.84DD122 pKa = 3.39LSVIRR127 pKa = 11.84GVLSLLSVFRR137 pKa = 11.84IMKK140 pKa = 9.95IPSTLKK146 pKa = 10.56LEE148 pKa = 4.94SITGPFTGLDD158 pKa = 3.39STLPKK163 pKa = 10.64YY164 pKa = 10.43EE165 pKa = 4.63LIKK168 pKa = 10.38IKK170 pKa = 10.37QVLPVFPQIKK180 pKa = 9.66PIRR183 pKa = 11.84LKK185 pKa = 10.71LLRR188 pKa = 11.84SAGPNCKK195 pKa = 10.08VSMLGIWLDD204 pKa = 3.35IKK206 pKa = 10.92AWSQHH211 pKa = 5.63SNFKK215 pKa = 9.68TLIEE219 pKa = 4.79FIQLFPNYY227 pKa = 10.25VEE229 pKa = 4.21FRR231 pKa = 11.84DD232 pKa = 3.74MLLQEE237 pKa = 4.94AYY239 pKa = 10.35NLRR242 pKa = 11.84LSVPSKK248 pKa = 10.52DD249 pKa = 3.3LHH251 pKa = 6.75LGKK254 pKa = 10.37LAIKK258 pKa = 10.28EE259 pKa = 3.98EE260 pKa = 4.33AAGKK264 pKa = 10.41ARR266 pKa = 11.84VFAITDD272 pKa = 3.99SITQSVMGPISDD284 pKa = 5.79AIFKK288 pKa = 8.81MLRR291 pKa = 11.84QIPMDD296 pKa = 3.32GTFNQSAPLDD306 pKa = 3.66RR307 pKa = 11.84LVQLSKK313 pKa = 11.16DD314 pKa = 3.4GAIAEE319 pKa = 4.42KK320 pKa = 10.9DD321 pKa = 3.71RR322 pKa = 11.84IFYY325 pKa = 10.75SYY327 pKa = 10.86DD328 pKa = 2.84LSAATDD334 pKa = 3.56RR335 pKa = 11.84LPINLQKK342 pKa = 10.85DD343 pKa = 3.3ILSIYY348 pKa = 9.5CGEE351 pKa = 4.22SFASKK356 pKa = 9.95WSILMTDD363 pKa = 3.87RR364 pKa = 11.84DD365 pKa = 3.63WYY367 pKa = 8.87LTKK370 pKa = 10.51EE371 pKa = 4.09KK372 pKa = 10.8RR373 pKa = 11.84NLRR376 pKa = 11.84YY377 pKa = 9.81SVGQPMGALSSWAMLALTHH396 pKa = 6.16HH397 pKa = 7.22FIVGMAANRR406 pKa = 11.84VGKK409 pKa = 10.47LGFNHH414 pKa = 6.26YY415 pKa = 10.85ALLGDD420 pKa = 5.56DD421 pKa = 4.49IVIADD426 pKa = 3.96KK427 pKa = 11.17SVADD431 pKa = 4.06SYY433 pKa = 11.42YY434 pKa = 10.37MIMTEE439 pKa = 3.8ILGVQINLSKK449 pKa = 11.1SLVSTNSFEE458 pKa = 4.1FAKK461 pKa = 10.7RR462 pKa = 11.84LVTLEE467 pKa = 4.31GEE469 pKa = 4.46VTPAGPANILLGLRR483 pKa = 11.84SLNGIPSIILDD494 pKa = 3.88MVNKK498 pKa = 9.33GVPVSEE504 pKa = 4.72EE505 pKa = 4.09SLEE508 pKa = 4.18SWMSTIPTVRR518 pKa = 11.84KK519 pKa = 9.46SQLEE523 pKa = 4.03KK524 pKa = 10.91VKK526 pKa = 10.18WVVKK530 pKa = 10.5GPFGFVPTAEE540 pKa = 4.32GLASFLTMSSSLTPVRR556 pKa = 11.84ANQIIHH562 pKa = 6.58AVRR565 pKa = 11.84YY566 pKa = 7.59TKK568 pKa = 10.03FQWDD572 pKa = 3.26LDD574 pKa = 3.55MWQKK578 pKa = 10.23VVKK581 pKa = 10.7DD582 pKa = 3.89NINYY586 pKa = 9.68RR587 pKa = 11.84ISIEE591 pKa = 3.88EE592 pKa = 4.35LYY594 pKa = 10.89FPVGFQDD601 pKa = 3.46YY602 pKa = 9.58PLDD605 pKa = 3.9FRR607 pKa = 11.84FSPIKK612 pKa = 10.1MEE614 pKa = 4.71IIKK617 pKa = 10.62SLSQDD622 pKa = 3.17LRR624 pKa = 11.84DD625 pKa = 4.42LGGQRR630 pKa = 11.84PTFRR634 pKa = 11.84LIFEE638 pKa = 4.73GPIIMFNYY646 pKa = 8.56YY647 pKa = 9.0RR648 pKa = 11.84QGYY651 pKa = 9.16ASEE654 pKa = 3.7IVNYY658 pKa = 9.88IKK660 pKa = 10.92YY661 pKa = 10.01LVHH664 pKa = 6.42QEE666 pKa = 4.07PEE668 pKa = 4.21FKK670 pKa = 10.31IGGSDD675 pKa = 3.24PFLPRR680 pKa = 11.84NFEE683 pKa = 4.05AFAFSSKK690 pKa = 10.5FKK692 pKa = 9.81GQKK695 pKa = 9.46FFEE698 pKa = 4.37QVRR701 pKa = 11.84EE702 pKa = 4.0HH703 pKa = 6.69LRR705 pKa = 11.84ANQLPMM711 pKa = 5.35
Molecular weight: 81.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S4GCV3|A0A0S4GCV3_9VIRU RNA dependent RNA polymerase OS=Sclerotinia sclerotiorum mitovirus 3 OX=1279099 GN=RdRp PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.03HH3 pKa = 6.45LKK5 pKa = 8.74LTTYY9 pKa = 10.56KK10 pKa = 10.76HH11 pKa = 6.66LDD13 pKa = 3.53LNINQADD20 pKa = 4.12RR21 pKa = 11.84FSNGAFAALSLKK33 pKa = 10.13LWYY36 pKa = 9.22PYY38 pKa = 10.55VRR40 pKa = 11.84LLIWSLRR47 pKa = 11.84LDD49 pKa = 3.78KK50 pKa = 10.92PSFMKK55 pKa = 10.34FATRR59 pKa = 11.84IEE61 pKa = 4.33SLWIHH66 pKa = 6.41NGLIFTVKK74 pKa = 10.19YY75 pKa = 10.55LKK77 pKa = 10.31EE78 pKa = 3.9CLRR81 pKa = 11.84ICQHH85 pKa = 5.93FVSGNPVLVTTEE97 pKa = 3.94MPISIVRR104 pKa = 11.84GIPTIIPGSIRR115 pKa = 11.84LQMHH119 pKa = 5.9SRR121 pKa = 11.84DD122 pKa = 3.39LSVIRR127 pKa = 11.84GVLSLLSVFRR137 pKa = 11.84IMKK140 pKa = 9.95IPSTLKK146 pKa = 10.56LEE148 pKa = 4.94SITGPFTGLDD158 pKa = 3.39STLPKK163 pKa = 10.64YY164 pKa = 10.43EE165 pKa = 4.63LIKK168 pKa = 10.38IKK170 pKa = 10.37QVLPVFPQIKK180 pKa = 9.66PIRR183 pKa = 11.84LKK185 pKa = 10.71LLRR188 pKa = 11.84SAGPNCKK195 pKa = 10.08VSMLGIWLDD204 pKa = 3.35IKK206 pKa = 10.92AWSQHH211 pKa = 5.63SNFKK215 pKa = 9.68TLIEE219 pKa = 4.79FIQLFPNYY227 pKa = 10.25VEE229 pKa = 4.21FRR231 pKa = 11.84DD232 pKa = 3.74MLLQEE237 pKa = 4.94AYY239 pKa = 10.35NLRR242 pKa = 11.84LSVPSKK248 pKa = 10.52DD249 pKa = 3.3LHH251 pKa = 6.75LGKK254 pKa = 10.37LAIKK258 pKa = 10.28EE259 pKa = 3.98EE260 pKa = 4.33AAGKK264 pKa = 10.41ARR266 pKa = 11.84VFAITDD272 pKa = 3.99SITQSVMGPISDD284 pKa = 5.79AIFKK288 pKa = 8.81MLRR291 pKa = 11.84QIPMDD296 pKa = 3.32GTFNQSAPLDD306 pKa = 3.66RR307 pKa = 11.84LVQLSKK313 pKa = 11.16DD314 pKa = 3.4GAIAEE319 pKa = 4.42KK320 pKa = 10.9DD321 pKa = 3.71RR322 pKa = 11.84IFYY325 pKa = 10.75SYY327 pKa = 10.86DD328 pKa = 2.84LSAATDD334 pKa = 3.56RR335 pKa = 11.84LPINLQKK342 pKa = 10.85DD343 pKa = 3.3ILSIYY348 pKa = 9.5CGEE351 pKa = 4.22SFASKK356 pKa = 9.95WSILMTDD363 pKa = 3.87RR364 pKa = 11.84DD365 pKa = 3.63WYY367 pKa = 8.87LTKK370 pKa = 10.51EE371 pKa = 4.09KK372 pKa = 10.8RR373 pKa = 11.84NLRR376 pKa = 11.84YY377 pKa = 9.81SVGQPMGALSSWAMLALTHH396 pKa = 6.16HH397 pKa = 7.22FIVGMAANRR406 pKa = 11.84VGKK409 pKa = 10.47LGFNHH414 pKa = 6.26YY415 pKa = 10.85ALLGDD420 pKa = 5.56DD421 pKa = 4.49IVIADD426 pKa = 3.96KK427 pKa = 11.17SVADD431 pKa = 4.06SYY433 pKa = 11.42YY434 pKa = 10.37MIMTEE439 pKa = 3.8ILGVQINLSKK449 pKa = 11.1SLVSTNSFEE458 pKa = 4.1FAKK461 pKa = 10.7RR462 pKa = 11.84LVTLEE467 pKa = 4.31GEE469 pKa = 4.46VTPAGPANILLGLRR483 pKa = 11.84SLNGIPSIILDD494 pKa = 3.88MVNKK498 pKa = 9.33GVPVSEE504 pKa = 4.72EE505 pKa = 4.09SLEE508 pKa = 4.18SWMSTIPTVRR518 pKa = 11.84KK519 pKa = 9.46SQLEE523 pKa = 4.03KK524 pKa = 10.91VKK526 pKa = 10.18WVVKK530 pKa = 10.5GPFGFVPTAEE540 pKa = 4.32GLASFLTMSSSLTPVRR556 pKa = 11.84ANQIIHH562 pKa = 6.58AVRR565 pKa = 11.84YY566 pKa = 7.59TKK568 pKa = 10.03FQWDD572 pKa = 3.26LDD574 pKa = 3.55MWQKK578 pKa = 10.23VVKK581 pKa = 10.7DD582 pKa = 3.89NINYY586 pKa = 9.68RR587 pKa = 11.84ISIEE591 pKa = 3.88EE592 pKa = 4.35LYY594 pKa = 10.89FPVGFQDD601 pKa = 3.46YY602 pKa = 9.58PLDD605 pKa = 3.9FRR607 pKa = 11.84FSPIKK612 pKa = 10.1MEE614 pKa = 4.71IIKK617 pKa = 10.62SLSQDD622 pKa = 3.17LRR624 pKa = 11.84DD625 pKa = 4.42LGGQRR630 pKa = 11.84PTFRR634 pKa = 11.84LIFEE638 pKa = 4.73GPIIMFNYY646 pKa = 8.56YY647 pKa = 9.0RR648 pKa = 11.84QGYY651 pKa = 9.16ASEE654 pKa = 3.7IVNYY658 pKa = 9.88IKK660 pKa = 10.92YY661 pKa = 10.01LVHH664 pKa = 6.42QEE666 pKa = 4.07PEE668 pKa = 4.21FKK670 pKa = 10.31IGGSDD675 pKa = 3.24PFLPRR680 pKa = 11.84NFEE683 pKa = 4.05AFAFSSKK690 pKa = 10.5FKK692 pKa = 9.81GQKK695 pKa = 9.46FFEE698 pKa = 4.37QVRR701 pKa = 11.84EE702 pKa = 4.0HH703 pKa = 6.69LRR705 pKa = 11.84ANQLPMM711 pKa = 5.35
MM1 pKa = 7.42KK2 pKa = 10.03HH3 pKa = 6.45LKK5 pKa = 8.74LTTYY9 pKa = 10.56KK10 pKa = 10.76HH11 pKa = 6.66LDD13 pKa = 3.53LNINQADD20 pKa = 4.12RR21 pKa = 11.84FSNGAFAALSLKK33 pKa = 10.13LWYY36 pKa = 9.22PYY38 pKa = 10.55VRR40 pKa = 11.84LLIWSLRR47 pKa = 11.84LDD49 pKa = 3.78KK50 pKa = 10.92PSFMKK55 pKa = 10.34FATRR59 pKa = 11.84IEE61 pKa = 4.33SLWIHH66 pKa = 6.41NGLIFTVKK74 pKa = 10.19YY75 pKa = 10.55LKK77 pKa = 10.31EE78 pKa = 3.9CLRR81 pKa = 11.84ICQHH85 pKa = 5.93FVSGNPVLVTTEE97 pKa = 3.94MPISIVRR104 pKa = 11.84GIPTIIPGSIRR115 pKa = 11.84LQMHH119 pKa = 5.9SRR121 pKa = 11.84DD122 pKa = 3.39LSVIRR127 pKa = 11.84GVLSLLSVFRR137 pKa = 11.84IMKK140 pKa = 9.95IPSTLKK146 pKa = 10.56LEE148 pKa = 4.94SITGPFTGLDD158 pKa = 3.39STLPKK163 pKa = 10.64YY164 pKa = 10.43EE165 pKa = 4.63LIKK168 pKa = 10.38IKK170 pKa = 10.37QVLPVFPQIKK180 pKa = 9.66PIRR183 pKa = 11.84LKK185 pKa = 10.71LLRR188 pKa = 11.84SAGPNCKK195 pKa = 10.08VSMLGIWLDD204 pKa = 3.35IKK206 pKa = 10.92AWSQHH211 pKa = 5.63SNFKK215 pKa = 9.68TLIEE219 pKa = 4.79FIQLFPNYY227 pKa = 10.25VEE229 pKa = 4.21FRR231 pKa = 11.84DD232 pKa = 3.74MLLQEE237 pKa = 4.94AYY239 pKa = 10.35NLRR242 pKa = 11.84LSVPSKK248 pKa = 10.52DD249 pKa = 3.3LHH251 pKa = 6.75LGKK254 pKa = 10.37LAIKK258 pKa = 10.28EE259 pKa = 3.98EE260 pKa = 4.33AAGKK264 pKa = 10.41ARR266 pKa = 11.84VFAITDD272 pKa = 3.99SITQSVMGPISDD284 pKa = 5.79AIFKK288 pKa = 8.81MLRR291 pKa = 11.84QIPMDD296 pKa = 3.32GTFNQSAPLDD306 pKa = 3.66RR307 pKa = 11.84LVQLSKK313 pKa = 11.16DD314 pKa = 3.4GAIAEE319 pKa = 4.42KK320 pKa = 10.9DD321 pKa = 3.71RR322 pKa = 11.84IFYY325 pKa = 10.75SYY327 pKa = 10.86DD328 pKa = 2.84LSAATDD334 pKa = 3.56RR335 pKa = 11.84LPINLQKK342 pKa = 10.85DD343 pKa = 3.3ILSIYY348 pKa = 9.5CGEE351 pKa = 4.22SFASKK356 pKa = 9.95WSILMTDD363 pKa = 3.87RR364 pKa = 11.84DD365 pKa = 3.63WYY367 pKa = 8.87LTKK370 pKa = 10.51EE371 pKa = 4.09KK372 pKa = 10.8RR373 pKa = 11.84NLRR376 pKa = 11.84YY377 pKa = 9.81SVGQPMGALSSWAMLALTHH396 pKa = 6.16HH397 pKa = 7.22FIVGMAANRR406 pKa = 11.84VGKK409 pKa = 10.47LGFNHH414 pKa = 6.26YY415 pKa = 10.85ALLGDD420 pKa = 5.56DD421 pKa = 4.49IVIADD426 pKa = 3.96KK427 pKa = 11.17SVADD431 pKa = 4.06SYY433 pKa = 11.42YY434 pKa = 10.37MIMTEE439 pKa = 3.8ILGVQINLSKK449 pKa = 11.1SLVSTNSFEE458 pKa = 4.1FAKK461 pKa = 10.7RR462 pKa = 11.84LVTLEE467 pKa = 4.31GEE469 pKa = 4.46VTPAGPANILLGLRR483 pKa = 11.84SLNGIPSIILDD494 pKa = 3.88MVNKK498 pKa = 9.33GVPVSEE504 pKa = 4.72EE505 pKa = 4.09SLEE508 pKa = 4.18SWMSTIPTVRR518 pKa = 11.84KK519 pKa = 9.46SQLEE523 pKa = 4.03KK524 pKa = 10.91VKK526 pKa = 10.18WVVKK530 pKa = 10.5GPFGFVPTAEE540 pKa = 4.32GLASFLTMSSSLTPVRR556 pKa = 11.84ANQIIHH562 pKa = 6.58AVRR565 pKa = 11.84YY566 pKa = 7.59TKK568 pKa = 10.03FQWDD572 pKa = 3.26LDD574 pKa = 3.55MWQKK578 pKa = 10.23VVKK581 pKa = 10.7DD582 pKa = 3.89NINYY586 pKa = 9.68RR587 pKa = 11.84ISIEE591 pKa = 3.88EE592 pKa = 4.35LYY594 pKa = 10.89FPVGFQDD601 pKa = 3.46YY602 pKa = 9.58PLDD605 pKa = 3.9FRR607 pKa = 11.84FSPIKK612 pKa = 10.1MEE614 pKa = 4.71IIKK617 pKa = 10.62SLSQDD622 pKa = 3.17LRR624 pKa = 11.84DD625 pKa = 4.42LGGQRR630 pKa = 11.84PTFRR634 pKa = 11.84LIFEE638 pKa = 4.73GPIIMFNYY646 pKa = 8.56YY647 pKa = 9.0RR648 pKa = 11.84QGYY651 pKa = 9.16ASEE654 pKa = 3.7IVNYY658 pKa = 9.88IKK660 pKa = 10.92YY661 pKa = 10.01LVHH664 pKa = 6.42QEE666 pKa = 4.07PEE668 pKa = 4.21FKK670 pKa = 10.31IGGSDD675 pKa = 3.24PFLPRR680 pKa = 11.84NFEE683 pKa = 4.05AFAFSSKK690 pKa = 10.5FKK692 pKa = 9.81GQKK695 pKa = 9.46FFEE698 pKa = 4.37QVRR701 pKa = 11.84EE702 pKa = 4.0HH703 pKa = 6.69LRR705 pKa = 11.84ANQLPMM711 pKa = 5.35
Molecular weight: 81.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
711 |
711 |
711 |
711.0 |
81.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.485 ± 0.0 | 0.563 ± 0.0 |
4.501 ± 0.0 | 4.501 ± 0.0 |
5.767 ± 0.0 | 5.485 ± 0.0 |
1.828 ± 0.0 | 8.579 ± 0.0 |
6.751 ± 0.0 | 11.814 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.235 ± 0.0 | 3.657 ± 0.0 |
5.204 ± 0.0 | 3.797 ± 0.0 |
5.204 ± 0.0 | 8.439 ± 0.0 |
4.219 ± 0.0 | 5.907 ± 0.0 |
1.688 ± 0.0 | 3.376 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
