
Halorubrum sp. E3
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halorubrum; unclassified Halorubrum
Average proteome isoelectric point is 4.89
Get precalculated fractions of proteins
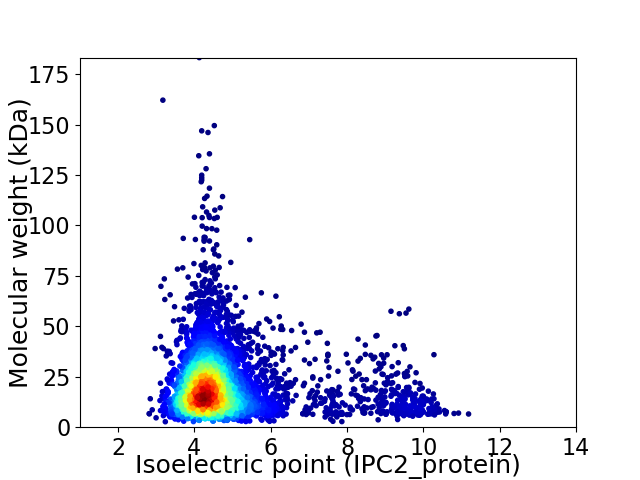
Virtual 2D-PAGE plot for 4984 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A256I9R1|A0A256I9R1_9EURY Phage repressor protein OS=Halorubrum sp. E3 OX=1483400 GN=DJ71_27740 PE=4 SV=1
MM1 pKa = 7.12SQQGVQEE8 pKa = 4.12EE9 pKa = 5.68LYY11 pKa = 10.68VDD13 pKa = 4.12QYY15 pKa = 11.52TLGLVGPDD23 pKa = 3.48QEE25 pKa = 4.57WAGTVADD32 pKa = 4.47GGTVTTYY39 pKa = 9.98TPPGCWGPMVTPSFRR54 pKa = 11.84GGHH57 pKa = 5.06EE58 pKa = 3.75VTRR61 pKa = 11.84PIRR64 pKa = 11.84VEE66 pKa = 3.78GAEE69 pKa = 4.01VGDD72 pKa = 3.82AVAIHH77 pKa = 6.26IRR79 pKa = 11.84DD80 pKa = 3.73VEE82 pKa = 4.51VTSMATSTGSMAEE95 pKa = 3.97RR96 pKa = 11.84EE97 pKa = 4.53GAFGDD102 pKa = 4.66DD103 pKa = 3.43PFVDD107 pKa = 4.29HH108 pKa = 7.33RR109 pKa = 11.84CPEE112 pKa = 4.74CGTTWPDD119 pKa = 3.24SVVEE123 pKa = 4.23GTGEE127 pKa = 3.82DD128 pKa = 3.76AIRR131 pKa = 11.84CAEE134 pKa = 4.18CGANASSFGFEE145 pKa = 3.79YY146 pKa = 10.41GYY148 pKa = 8.93TVAFDD153 pKa = 3.68HH154 pKa = 6.64EE155 pKa = 4.52NAVGITLDD163 pKa = 3.33EE164 pKa = 5.25DD165 pKa = 4.29GAHH168 pKa = 6.6EE169 pKa = 4.35LALDD173 pKa = 4.01ADD175 pKa = 4.34EE176 pKa = 5.95VMDD179 pKa = 3.57IPEE182 pKa = 4.49NARR185 pKa = 11.84QHH187 pKa = 6.73PILLYY192 pKa = 10.55EE193 pKa = 4.45PDD195 pKa = 4.17GMPGTLGRR203 pKa = 11.84LRR205 pKa = 11.84PFIGNIGTTPPVTMPDD221 pKa = 2.99SHH223 pKa = 6.53NAGDD227 pKa = 4.37FGQNLIGADD236 pKa = 3.19HH237 pKa = 7.78DD238 pKa = 4.6YY239 pKa = 11.38GVEE242 pKa = 4.07TEE244 pKa = 4.48EE245 pKa = 5.77DD246 pKa = 3.45LEE248 pKa = 4.19QRR250 pKa = 11.84TDD252 pKa = 2.72GHH254 pKa = 6.05MDD256 pKa = 3.26VPEE259 pKa = 4.21VRR261 pKa = 11.84AGATLICPVVVDD273 pKa = 3.75GGGIYY278 pKa = 10.68VGDD281 pKa = 3.75LHH283 pKa = 7.3ANQGDD288 pKa = 4.47GEE290 pKa = 4.6LSLHH294 pKa = 5.12TTDD297 pKa = 3.8VSGTVTMDD305 pKa = 3.09VEE307 pKa = 4.55VIEE310 pKa = 5.8DD311 pKa = 3.54VDD313 pKa = 4.45LDD315 pKa = 4.47GPVLLPNEE323 pKa = 4.07EE324 pKa = 4.42DD325 pKa = 3.98LPFISAPYY333 pKa = 9.75SDD335 pKa = 4.78EE336 pKa = 3.89EE337 pKa = 4.43VAAGDD342 pKa = 4.45DD343 pKa = 3.6LGAEE347 pKa = 4.38HH348 pKa = 6.89GVDD351 pKa = 3.44VV352 pKa = 4.67
MM1 pKa = 7.12SQQGVQEE8 pKa = 4.12EE9 pKa = 5.68LYY11 pKa = 10.68VDD13 pKa = 4.12QYY15 pKa = 11.52TLGLVGPDD23 pKa = 3.48QEE25 pKa = 4.57WAGTVADD32 pKa = 4.47GGTVTTYY39 pKa = 9.98TPPGCWGPMVTPSFRR54 pKa = 11.84GGHH57 pKa = 5.06EE58 pKa = 3.75VTRR61 pKa = 11.84PIRR64 pKa = 11.84VEE66 pKa = 3.78GAEE69 pKa = 4.01VGDD72 pKa = 3.82AVAIHH77 pKa = 6.26IRR79 pKa = 11.84DD80 pKa = 3.73VEE82 pKa = 4.51VTSMATSTGSMAEE95 pKa = 3.97RR96 pKa = 11.84EE97 pKa = 4.53GAFGDD102 pKa = 4.66DD103 pKa = 3.43PFVDD107 pKa = 4.29HH108 pKa = 7.33RR109 pKa = 11.84CPEE112 pKa = 4.74CGTTWPDD119 pKa = 3.24SVVEE123 pKa = 4.23GTGEE127 pKa = 3.82DD128 pKa = 3.76AIRR131 pKa = 11.84CAEE134 pKa = 4.18CGANASSFGFEE145 pKa = 3.79YY146 pKa = 10.41GYY148 pKa = 8.93TVAFDD153 pKa = 3.68HH154 pKa = 6.64EE155 pKa = 4.52NAVGITLDD163 pKa = 3.33EE164 pKa = 5.25DD165 pKa = 4.29GAHH168 pKa = 6.6EE169 pKa = 4.35LALDD173 pKa = 4.01ADD175 pKa = 4.34EE176 pKa = 5.95VMDD179 pKa = 3.57IPEE182 pKa = 4.49NARR185 pKa = 11.84QHH187 pKa = 6.73PILLYY192 pKa = 10.55EE193 pKa = 4.45PDD195 pKa = 4.17GMPGTLGRR203 pKa = 11.84LRR205 pKa = 11.84PFIGNIGTTPPVTMPDD221 pKa = 2.99SHH223 pKa = 6.53NAGDD227 pKa = 4.37FGQNLIGADD236 pKa = 3.19HH237 pKa = 7.78DD238 pKa = 4.6YY239 pKa = 11.38GVEE242 pKa = 4.07TEE244 pKa = 4.48EE245 pKa = 5.77DD246 pKa = 3.45LEE248 pKa = 4.19QRR250 pKa = 11.84TDD252 pKa = 2.72GHH254 pKa = 6.05MDD256 pKa = 3.26VPEE259 pKa = 4.21VRR261 pKa = 11.84AGATLICPVVVDD273 pKa = 3.75GGGIYY278 pKa = 10.68VGDD281 pKa = 3.75LHH283 pKa = 7.3ANQGDD288 pKa = 4.47GEE290 pKa = 4.6LSLHH294 pKa = 5.12TTDD297 pKa = 3.8VSGTVTMDD305 pKa = 3.09VEE307 pKa = 4.55VIEE310 pKa = 5.8DD311 pKa = 3.54VDD313 pKa = 4.45LDD315 pKa = 4.47GPVLLPNEE323 pKa = 4.07EE324 pKa = 4.42DD325 pKa = 3.98LPFISAPYY333 pKa = 9.75SDD335 pKa = 4.78EE336 pKa = 3.89EE337 pKa = 4.43VAAGDD342 pKa = 4.45DD343 pKa = 3.6LGAEE347 pKa = 4.38HH348 pKa = 6.89GVDD351 pKa = 3.44VV352 pKa = 4.67
Molecular weight: 37.29 kDa
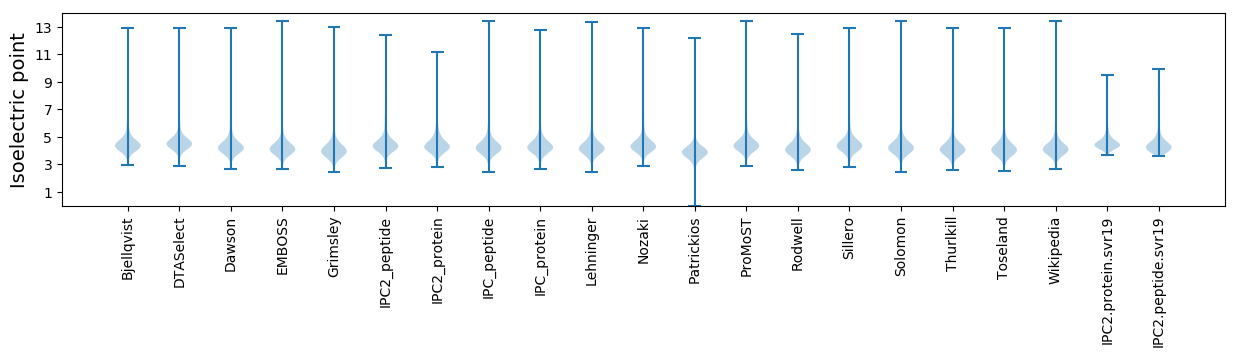
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A256I5Q1|A0A256I5Q1_9EURY S-adenosylmethionine synthetase (Fragment) OS=Halorubrum sp. E3 OX=1483400 GN=DJ71_28470 PE=4 SV=1
MM1 pKa = 7.62AATSGPLASLARR13 pKa = 11.84SRR15 pKa = 11.84SSVIRR20 pKa = 11.84SSLSPRR26 pKa = 11.84FALAASGRR34 pKa = 11.84SRR36 pKa = 11.84ATTALRR42 pKa = 11.84FASRR46 pKa = 11.84LIVSARR52 pKa = 11.84ALVSAMRR59 pKa = 11.84AANAPP64 pKa = 3.54
MM1 pKa = 7.62AATSGPLASLARR13 pKa = 11.84SRR15 pKa = 11.84SSVIRR20 pKa = 11.84SSLSPRR26 pKa = 11.84FALAASGRR34 pKa = 11.84SRR36 pKa = 11.84ATTALRR42 pKa = 11.84FASRR46 pKa = 11.84LIVSARR52 pKa = 11.84ALVSAMRR59 pKa = 11.84AANAPP64 pKa = 3.54
Molecular weight: 6.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1007089 |
24 |
1668 |
202.1 |
21.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.051 ± 0.061 | 0.692 ± 0.014 |
8.838 ± 0.047 | 8.581 ± 0.056 |
3.274 ± 0.028 | 8.595 ± 0.044 |
1.948 ± 0.017 | 4.108 ± 0.029 |
1.949 ± 0.027 | 8.64 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.696 ± 0.018 | 2.388 ± 0.026 |
4.67 ± 0.023 | 2.403 ± 0.021 |
6.64 ± 0.047 | 5.524 ± 0.03 |
6.24 ± 0.033 | 8.893 ± 0.041 |
1.129 ± 0.016 | 2.742 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
